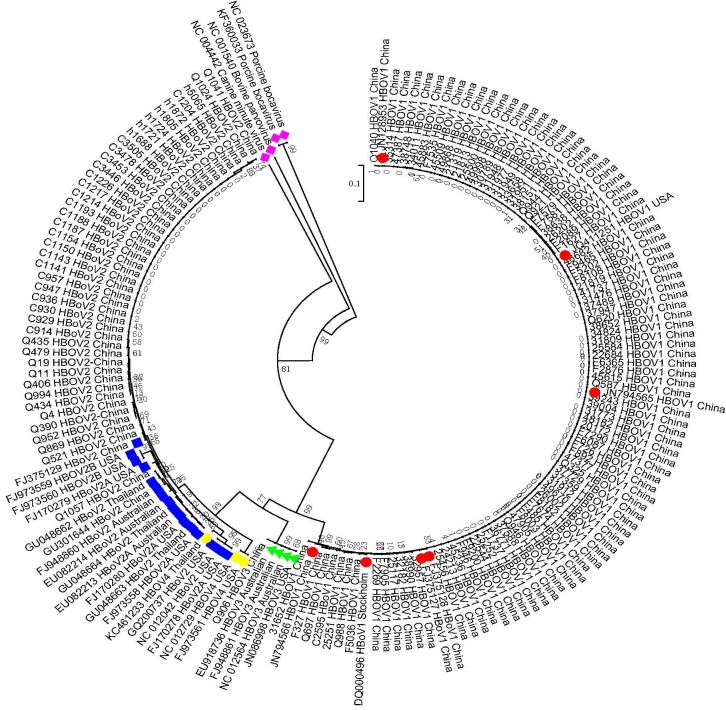
Fig 1. NP1/VP1 gene sequences (n = 131) obtained from clinical samples and GenBank sequences for HBoV1-4 were used for tree construction.
The phylogenetic tree was constructed using the neighbor-joining method based on the Kimura -parameter model with 1,000 bootstrap replicates, using the MEGA 6.0 software program. A red dot denotes an HBoV1 strain downloaded from GenBank, a blue box denotes an HBoV2 sequence from GenBank, a green triangle denotes an HBoV3 sequence from GenBank, a yellow diamond denotes an HBOV4 sequence from GenBank, and a purple diamond denotes other parvoviruses. All remaining accession numbers represent the 131 sequences obtained from clinical samples in the present study.