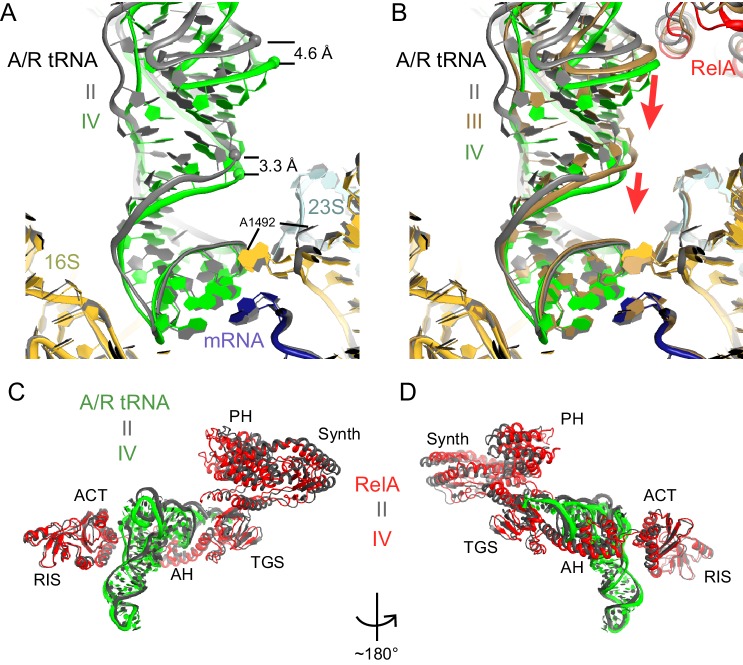
Figure 5. A/R tRNA and RelA rearrange toward the 30S subunit in Structures II to IV.
(A) A/R tRNA settles into the decoding center of the 30S subunit between Structures II (grey) and IV (colored as in Figure 1). Structures II and IV were aligned on the 16S rRNA. RelA is not shown. The positions of A1492 in Structures II and IV are labeled for reference. (B) A/R tRNA and RelA positions in Structures II (grey), III (gold) and IV (colored as in Figure 1). (C) and (D) Two views showing that RelA shifts with the A/R tRNA between Structure II (grey) and Structure IV (colored as in Figure 1). The TGS domain, which interacts with the acceptor arm of A/R tRNA, moves more than the RIS and ACT domains. The superposition of Structures II and IV was performed by structural alignment of the 16S rRNA.