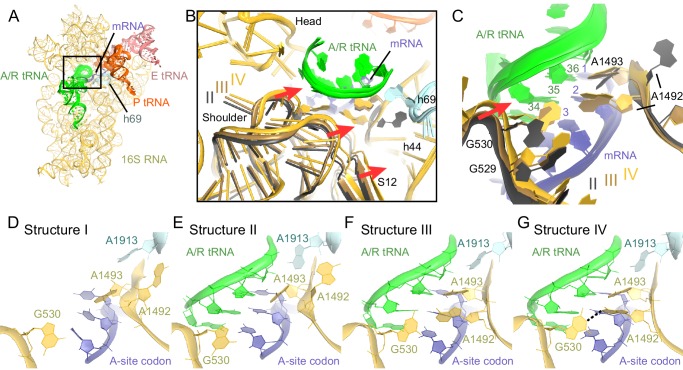
Figure 6. Closure of the 30S subunit and decoding-center rearrangements in Structures II, III and IV.
(A) A view down on the 30S subunit from the inter-subunit interface shows the position of the decoding center (boxed). The 50S subunit (except for helix 69), small ribosomal proteins and RelA are omitted for clarity. (B) The conformational differences in the 30S subunits of Structures II, III and IV suggest a domain-closure pathway. From Structure II to IV, the 30S shoulder is shifted by more than 4 Å toward the 30S head. The superposition of Structures II, III, and IV was performed by structural alignment of nt 980–1200 of 16S rRNA, corresponding to the 30S head. (C) Conformational differences in the decoding-center’s universally conserved nucleotides A1492, A1493, and G530 of Structure II, III, and IV are shown after alignment as in (B). (D) The decoding center of Structure I, which lacks A/R tRNA, is similar to that of the 30S domain-open structures with a vacant A site (Ogle et al., 2001; Jenner et al., 2010). (E) The decoding center of Structure II reveals a previously unseen state, in which the domain-open 30S subunit contains a tRNA in the A site. A1493 is near the first base pair of the codon-anticodon helix; A1492 is in helix 44, whereas G530 adopts the conformation previously observed in the absence of the A-site tRNA (Ogle et al., 2001; Jenner et al., 2010). (F) The decoding center of Structure III reveals a previously unseen state, in which the 30S subunit adopts an intermediate domain-closure conformation. A1493 and A1492 interact with the first and second base pairs of the codon-anticodon helix, respectively, whereas G530 is oriented toward A1492. (G) The decoding center of Structure IV, with a closed 30S conformation, comprises A1493 and A1492 forming A-minor interactions with the first two base pairs of the codon-anticodon helix, whereas G530 is shifted toward helix 44 and interacts with A1492. This conformation resembles that of other 30S domain-closed structures in pre-accommodation-like 70S•EF-Tu•aa-tRNA complexes (Stark et al., 2001; Valle, 2002; Schmeing et al., 2009) and 70S complexes with fully accommodated A/A tRNA (Voorhees et al., 2009; Jenner et al., 2010; Demeshkina et al., 2012). Proteins are omitted for clarity in (C–G).
DOI: http://dx.doi.org/10.7554/eLife.17029.019
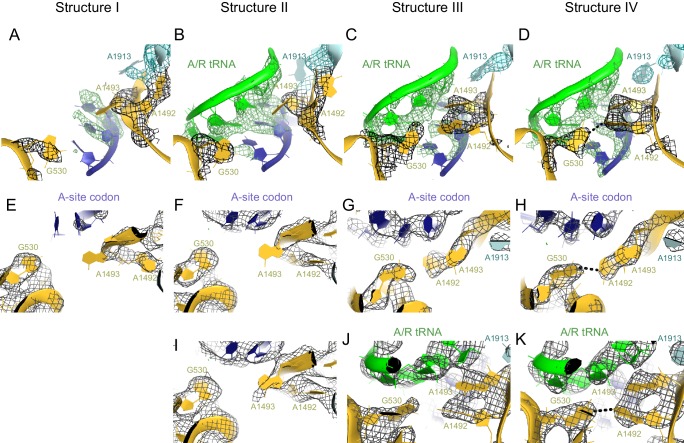
Figure 6—figure supplement 1. Comparison of the 30S subunits of Structures II, III and IV reveals domain closure of the 30S subunit from Structure II to IV.