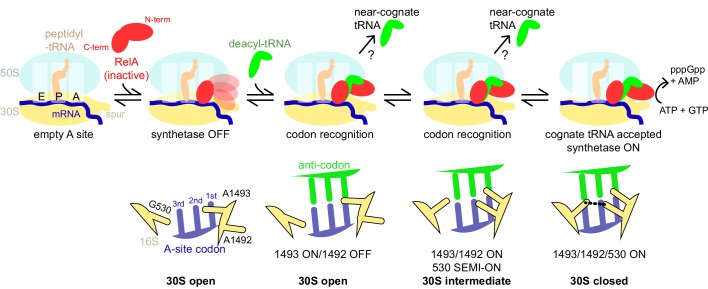
Figure 7. Schematic of the mechanism of RelA activation by the ribosome and cognate deacyl-tRNA.
The C-terminal domains of RelA, the RIS and the ACT, bind the ribosome at the intersubunit bridge B1a near the vacant A site, but the synthetase remains unbound and inactive. When deacyl-tRNA binds to the ribosomal A site, the decoding center controls the selection of cognate tRNA, coupled with domain closure of the 30S subunit. The codon recognition checkpoints are mediated by distinct positions of the universally conserved nucleotides of the decoding center A1492, A1493 and G530. Upon binding of cognate deacyl-tRNA to the ribosome, the RelA synthetase domain is exposed in the vicinity of the 30S spur and is activated for (p)ppGpp synthesis by alleviation of RelA autoinhibition and interactions with the ribosome.