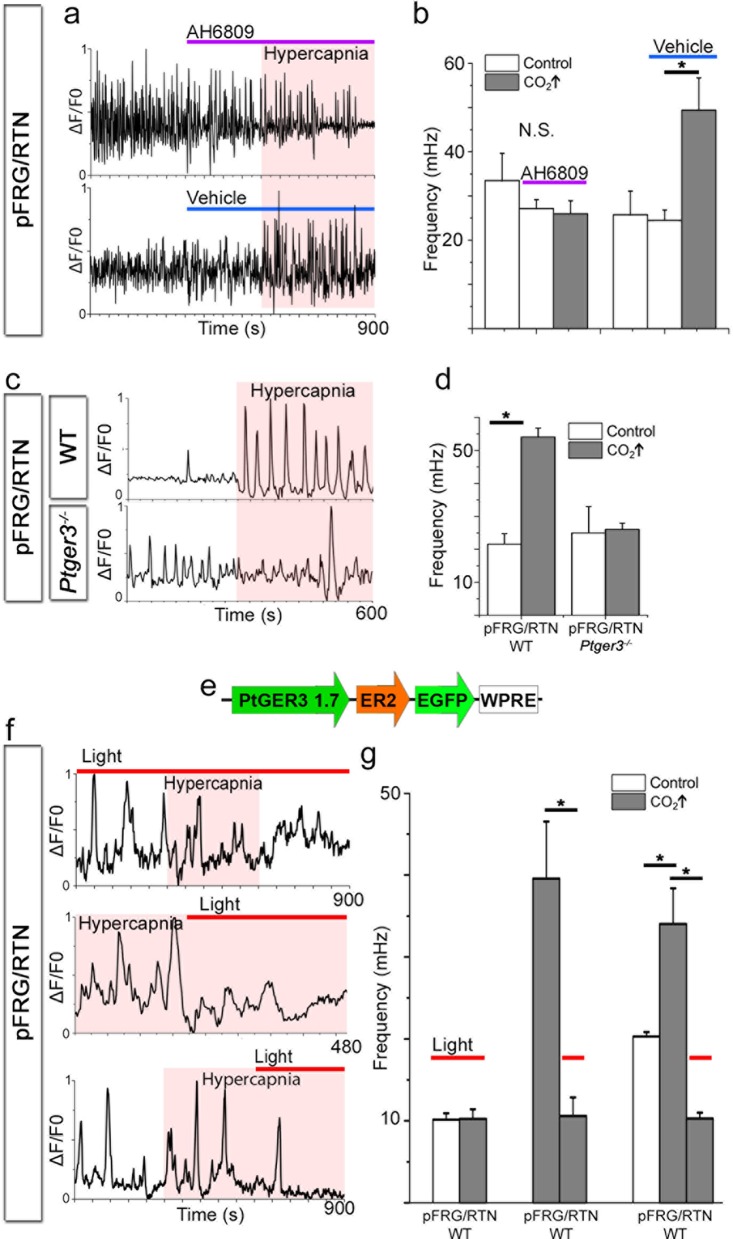
Figure 13. PGE2, acting through EP3R, is crucial for the hypercapnic response.
Pharmacological inhibition of EP3R by the EP receptor antagonist AH6809 inhibited the response to hypercapnia (increased pCO2[CO2↑]) in the pFRG/RTN (N=6, n=472, N.S.; a–b). The hypercapnic response was also absent in pFRG/RTN slices lacking EP3R (Ptger3−/−; N=5, n=348, N.S.; c–d). Layout of the lentivirus containing Halo57 (ER2) and eGFP genes under the control of the EP3R promoter (Ptger3) used for optogenetics (WPRE=gene enhancing element; e). During optogenetic silencing of Ptger3-expressing cells, no frequency changes were observed in response to hypercapnia (f, top trace; g, left graph). The hypercapnic response was also reversed by activating Ptger3-Halo57 (f, middle and bottom trace; g, middle and right graph). Red line: Halo57 activation in response to 625 nm light. N: slices, n: cells. Data are presented as means ± SD. *p<0.05. Source data are available in a separate source data file.
DOI: http://dx.doi.org/10.7554/eLife.14170.058