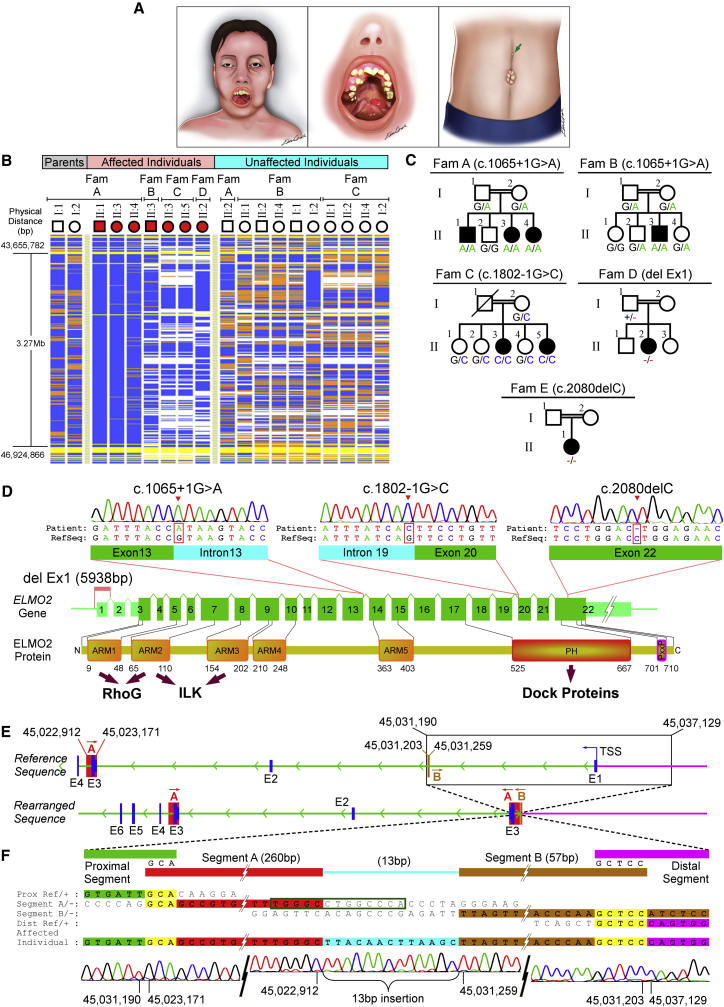
Figure 1.
Identification of the ELMO2 Mutations that Cause VMOS
(A) Illustration of the morphological VMOS findings. Maxillomandibular enlargement is evident, both extraorally and intraorally. Facial asymmetry due to bone enlargement is shown on the left. Exophthalmos and loss of vision usually accompany the disease in later stages. Ectopic eruption, impaction, and displacement of the teeth, as well as severe alveolar bone expansion are common findings in intraoral examination (middle). Supraumbilical raphe (green arrow) and umbilical hernia are the primary extraosseous findings in VMOS (right).
(B) Homozygosity mapping via VIGENOS showed a 3.27 Mbp candidate region in chromosome 20q13. Homozygous genotypes identical to the genotype data obtained from the affected individual A-II:1 are shown in blue. Contrasting homozygote genotypes are shown in white, whereas heterozygous genotypes appear in orange. Non-informative genotypes resulting from heterozygous SNPs in parent-child trios are shown in yellow. Note that all affected individuals are homozygous for the candidate region; however, each of the four families have a different haplotype for this interval.
(C) Pedigrees of families A–E. Mutations found in ELMO2 are shown for each family and the genotypes for the corresponding mutation are indicated below each individual whose DNA samples were available.
(D) Schematic representation of homozygous ELMO2 mutations co-segregating with VMOS in the five families. ELMO2 contains a total of 22 exons (green boxes). The untranslated regions of the exons are denoted with smaller light green boxes. In families A and B, the c.1065+1G>A mutation substitutes the first nucleotide of the 13th intron in the splice donor site. In family C, the c.1802−1G>C mutation substitutes the last nucleotide of the 19th intron in the splice acceptor site. In family D, a complex rearrangement involving a 5,938 bp deletion removes the first exon of ELMO2. In family E, the c.2080delC mutation deletes one cytosine, which leads to a frameshift predicted to produce a longer protein. Below is a schematic representation of the ELMO2. ELMO2 protein domains are linked by arrows to their interacting proteins. Abbreviations are as follows: ARM, Armadillo repeat; PH, pleckstrin homology domain; PxxP, proline-rich motif.
(E) Complex rearrangement schematized on the genomic sequence. The upper line illustrates the 5′ end of wild-type ELMO2. The exons are shown as blue boxes, and segments A and B, which represent inserted segments in the complex rearrangement (see Figure S1A), are shown as red and light brown boxes, respectively. The green arrowheads indicate the direction of transcription. The black rectangle shows the deleted portion of the genome. Abbreviation: TSS, transcription start site. The bottom line shows the rearranged sequence. Note that inserted sequences A and B are in reverse orientation (inverted). Genomic positions are indicated where applicable.
(F) Enlarged view of the complex rearrangement region. The upper part represents the sequence alignment of the breakpoint junctions, showing the homology with four genomic regions, namely proximal segment (green) (centromeric end of the breakpoint), inverted segment A (red), inverted segment B (light brown), and distal segment (purple) (telomeric end of the breakpoint). Two of the breakpoint junctions share 3-bp- and 5-bp-long microhomology sequences (shown in yellow), whereas a 13-bp-long joining segment shown in blue joins segments A and B. Nucleotide sequences homologous to RefSeq are depicted in bold. The green box is a 13-bp-long inverted repeat sequence at the breakpoint junction of segment A. The lower part shows the corresponding Sanger sequences for the three breakpoint junctions. The genomic positions of the nucleotides in RefSeq are indicated below the electropherogram.
Also see Figure S2 for details.