Figure 3.
Location and Structural Modeling of Identified RCBTB1 Missense Variants
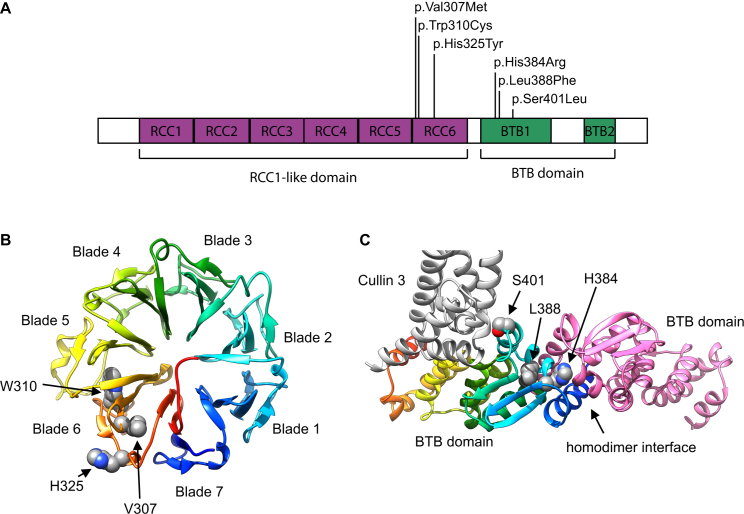
(A) Schematic diagram of RCBTB1 shows the location of the missense variants within two distinct domains: three (F1–F4) in the sixth repeat of the RCC1-like domain (RLD), i.e., RCC6, and two (F5 and F6) in the first BTB domain (BTB1).
(B) A homology model for the β-propeller structure of the RLD is shown in rainbow colors, evolving from blue (N-terminal) to red (C-terminal). The RCBTB1 RLD contains seven repeats that form a seven-bladed β-propeller, in which each blade consists of a four-stranded antiparallel β sheet. The p.Val307Met (c.919G>A), p.Trp310Cys (c.930G>T), and p.His325Tyr (c.973C>T) variants are all found in the sixth blade. Val307 and Trp310 are part of the third strand of blade 6. Trp310 is a conserved aromatic. The bulky indole group of Trp310 is buried in the hydrophobic core between blades five and six and makes extensive Van der Waals contacts with three aliphatic sidechains of blade five. p.Trp310Cys therefore introduces a big void between both blades and is probably highly destabilizing. Val307 is part of a hydrophobic core between blades six and seven. The more bulky methionine side chain introduced by p.Val307Met is clashing with residues of blade seven. His325 cannot be modeled accurately because alignments with different methods and against different templates give diverging outcomes for the exact position of this residue. Most likely, His325 is surface exposed at the end of the fourth strand of blade six.
(C) A homology model for the RCBTB1 BTB domain is shown in rainbow colors, evolving from blue (N-terminal) to red (C-terminal). Another interacting RCBTB1 BTB domain is shown in pink. Part of an interacting CUL3 molecule is shown in gray. His384 is found at the BTB homodimerization interface. A histidine is present at this position in nine of ten BTB structures aligned with the RCBTB1 BTB domain.8 His384 forms an extensive hydrogen-bond network at the interfacial area and makes a direct Van der Waals contact with the homodimerization partner. p.His384Arg (c.1151A>G) disrupts the hydrogen-bonding network, and accommodation of two bulky arginine residues in the homodimer interface is impossible. Leu388 is an extremely conserved residue of the hydrophobic BTB core and is identical in all ten crystal structures. p.Leu388Phe (c.1164G>T) introduces drastic steric clashes of the phenyl ring with surrounding hydrophobic residues. Ser401 is a surface-exposed residue that is either close to or at the edge of the BTB-CUL3 interface, depending on the template that was used. It is unclear whether p.Ser401Leu (c.1202C>T) can disrupt the interaction with CUL3. Homology models were built on the basis of different structure templates with YASARA Structure.9, 10 Additional models were built in MODELER and YASARA Structure with alignments based on HHPRED and Phyre2.8, 9, 10, 11, 12 On the basis of the initial models, the alignments were edited for model improvement, as judged by the DOPE score in MODELER, visual inspection, and Verify_3D 3D profile analysis of the models.11, 13 The models are based on template structures of RLD (PDB: 4O2W) and the BTB domain complex (PDB: 4J8Z and 4AP2). The effects of the variants were analyzed in YASARA Structure.9, 10 Figures were generated with UCSF Chimera.14