Figure 1.
Recessive CIT Variants in Microlissencephaly
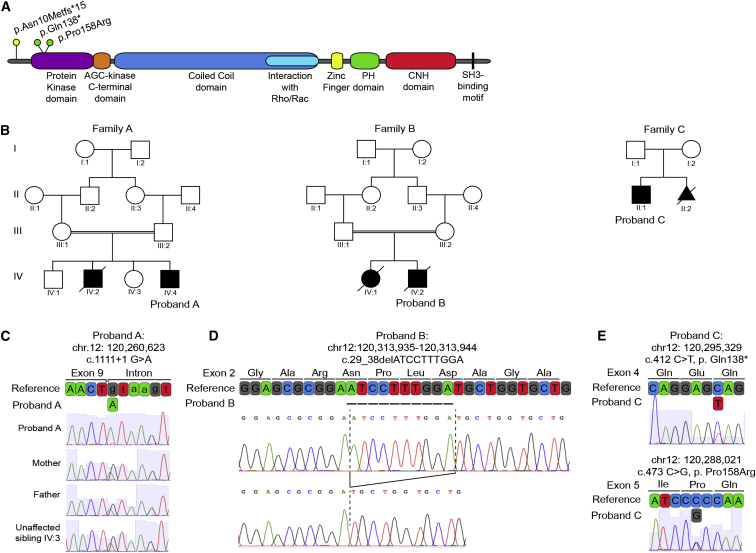
(A) Domain organization of CIT shows pathogenic coding variants in relation to protein domains.
(B) Pedigrees of the families investigated; probands are noted.
(C) Chromatographs and schematic of the boundary between exon 9 and intron 9 of WT reference CIT, the homozygous splice donor variant from proband A (c.1111+1G>A), and the cryptic splice donor site four bases downstream. Parents and unaffected sibling IV:3 are heterozygous carriers.
(D) Chromatographs defining the homozygous 10 bp CIT exon 2 deletion (c.29_38delATCCTTTGGA; hg19 chr12: 120,313,935–120,313,944) amplified from proband B. This deletion creates a frameshift leading to a premature stop 15 codons downstream (not shown).
(E) Chromatographs and schematic of reference CIT exons 4 and 5 and the corresponding nonsense (c.412C>T [p.Gln138∗]) and missense (c.473C>G [p.Pro158Arg]) variants identified in proband C.