Figure 3.
Functional Analysis of CIT c.1111+1G>A Confirms Its Negative Impact on RNA Splicing
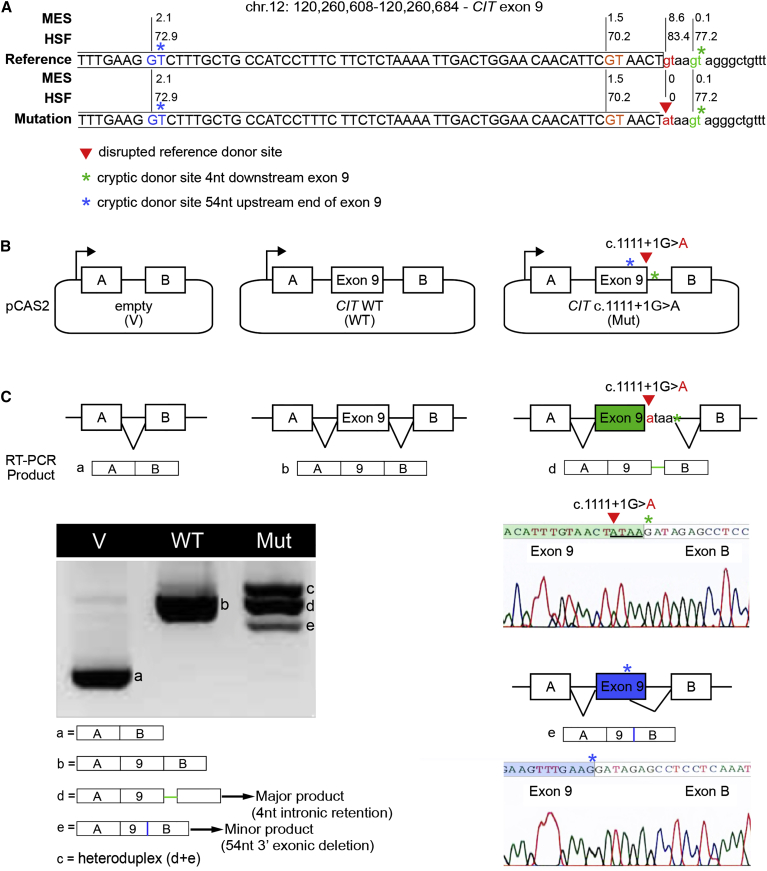
(A) Schematic representation of CIT sequence across the boundary between exon 9 and intron 9. The reference donor site (WT, red nucleotides) and cryptic donor sites (blue, orange, and green nucleotides) were delineated and scored with MaxEntScan (MES) and Human Splicing Finder (HSF) as shown in Figure S1. Only splice sites predicted by both algorithms in the WT sequence are shown. Exonic and intronic sequences are indicated by upper and lower case, respectively. The CIT c.1111+1G>A mutation from proband A (red arrowhead) disrupts the WT donor site. Asterisks indicate cryptic splice sites activated in the presence of c.1111+1G>A.
(B) A minigene splicing assay was performed as previously described. The structures of the CIT exon 9 minigenes were used in the splicing reporter assay. Arrows represent the CMV promoter. Boxes represent exons, and lines in between indicate introns.
(C) Analysis of WT and mutant (Mut) minigene splicing patterns. Transcripts were analyzed by RT-PCR after expression in HeLa cells with the use of primers specific to the minigene’s exons A and B (Table S1). RT-PCR products were separated on an agarose gel and sequenced. Aberrantly spliced products, accompanied by chromatographs, are illustrated on the right.