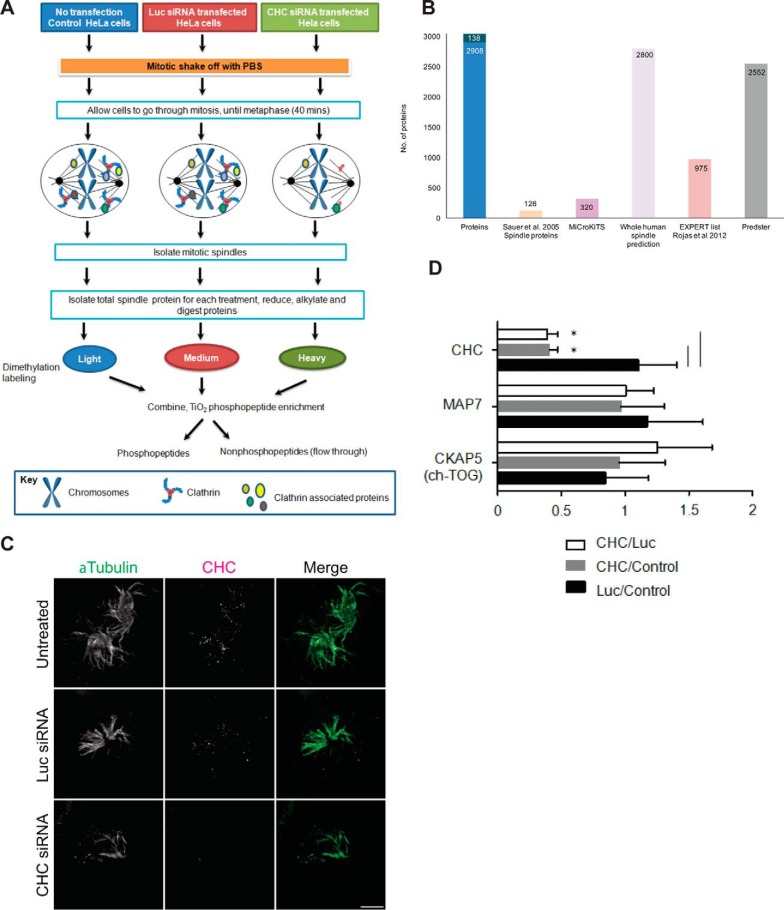
Fig. 1.
Mitotic spindle proteomics. A, Untransfected HeLa S3 cells and cells transfected with Luc-targeting siRNA or CHC-targeting siRNA were synchronized to pro-metaphase with Nocodazole and collected by mitotic shake-off. The workflow describes isolation of mitotic spindle proteins, tryptic digestion, dimethylation labeling of tryptic peptides, followed by preparation of each sample for analysis of phosphopeptides and nonphosphopeptides on the Orbitrap Velos mass spectrometer. B, Bar graph depicting the number of proteins that overlap between proteins identified in our study (Proteins) and proteins classified as localizing to the spindle by Sauer et al., 2005 (Sauer spindle list) (3), MiCroKiTS, predicted spindle proteins in the whole human proteome (SPIP), the Expert list (37) and the online tool Predster. C, Representative wide-field microscopy images of isolated mitotic spindles. Spindle structures of untreated cells and cells treated with siRNA against Luc or CHC stained for α-Tubulin (green) and CHC (magenta). Scale bar: 5 μm. D, Graph shows the relative change in abundance (mean ± S.E.) of clathrin, the microtubule- associated protein Map7 and ch-TOG by quantitative mass spectrometry. n≥3. *, p < 0.05.