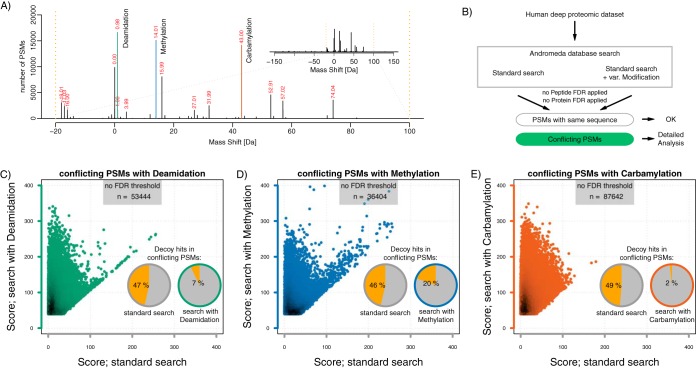
Fig. 1.
Modified peptides as a source of false spectra identification. A, Distribution of most frequent mass shifts of dependent peptides in the region from −20 to 100 Da. A view on the wider mass region is shown in the insert B, Raw data files were analyzed without (standard search) or with additional selected variable modifications: deamidation (Gln, Asn), methylation (Lys, Glu, Asp) or carbamylation (peptide N-term). All searches included methionine oxidation and protein N-terminal acetylation as variable modifications. C–E, Pairwise comparison of Andromeda scores from conflicting PSMs in a search with and without deamidation, methylation or carbamylation, as indicated. The fraction of decoy database matches among conflicting PSMs is given for either search variant in the pie charts.