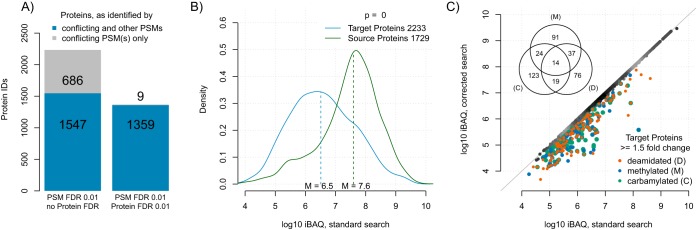
Fig. 3.
Modified peptides as a source of false protein identification and quantification. A, Protein identifications because of misidentified modified peptides in a standard search. The number of false identifications is given at a PSM FDR cut-off at 0.01 with and without additional protein FDR cut-off at 0.01. B, Density plot of protein log10 iBAQ values as estimates of protein abundances. Source proteins of modified peptides and proteins that were misidentified by modified peptides (target proteins) are depicted. Dashed lines indicate the median of the respective distribution. C, Pairwise comparison of abundance estimates of target proteins in a standard search and a search where conflicting PSMs were removed (corrected). Target proteins that were overestimated by modified peptides ≥1.5-fold-change are highlighted as indicated. Decoy database hits were removed for all subfigures. A cut-off at PSM FDR 0.01 and no protein FDR-threshold was applied for subfigures b and c.