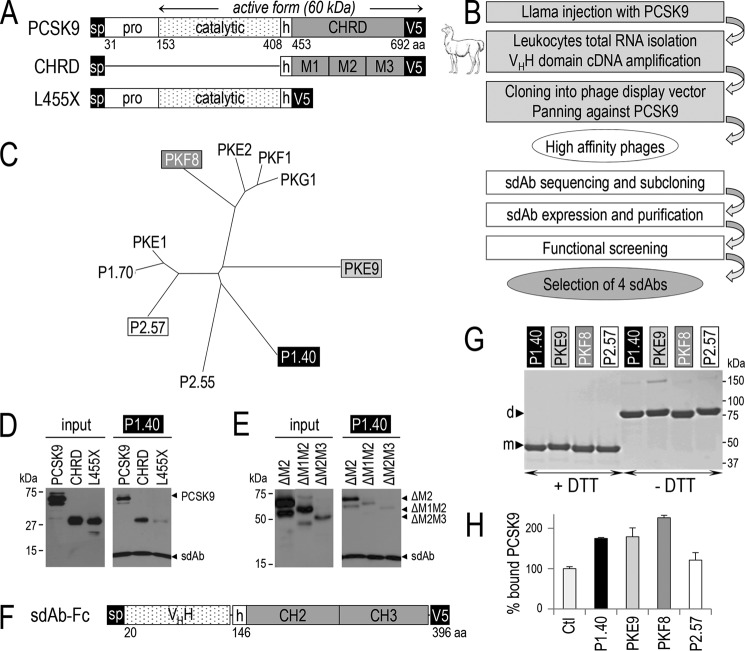
FIGURE 1.
Generation of llama PCSK9-specific sdAbs, PCSK9 binding to sdAb, effect of the sdAb on LDLR binding. The primary structure of PCSK9 and its deletants are shown: aa, amino acid; sp, signal peptide; pro, prodomain; h, hinge domain; CHRD, Cys/His-rich domain and its modules M1, M2, and M3 (A). Flow chart of the steps for llama sdAb selection (B). Phylogenetic tree deduced from the alignment of the full protein sequences of 10 selected sdAbs (C). The media of HEK293 transiently expressing PCSK9 or its deletants lacking either the pro- and catalytic domains (CHRD) or the CHRD (L455X) and the sdAb P1.40 were mixed and immunoprecipitated with TALON metal affinity resin. The input (conditioned media containing the different PCSK9 variants) and pellets were analyzed by WB with mAb-V5 antibody after SDS-PAGE on 12% Tris-glycine gels (D). Similar experiments were performed on PCSK9 variants lacking the CHRD modules M2 alone (ΔM2), M2 and M1 (ΔM1M2) or M2 and M3 (ΔM2M3) (E). D and E are representative of three independent experiments. Schematic of the representative fusion of P1.40 with a mouse Fc comprising the hinge (h), CH2, and CH3 domains (F). Purified sdAb-Fcs (2 μg) were separated by SDS-PAGE in the presence (+) or absence (−) of DTT and revealed by Coomassie staining (G). The four sdAb-Fcs were incubated with PCSK9 and tested for their ability to prevent binding of PCSK9 to the LDLR EGF-AB coated on a binding assay plate (H). This assay is an average of three technical replicates.