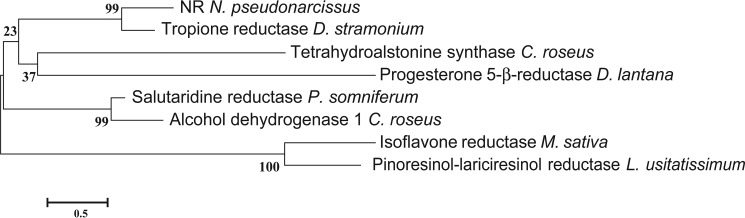
FIGURE 2.
Phylogenetic relationship of NR to other plant reductases and dehydrogenases. Amino acid sequences of all of the enzymes were taken from NCBI (PDB 2AE2, Q071NO.1, AJO70763.1, AKF02528.1, AAC48976.1, ABW24501.1, and AAS76634.1). The phylogenetic tree was constructed using MEGA (43), and the amino acid sequences were aligned using MUSCLE. The LG model was used to construct a Maximum Likelihood tree using a bootstrap replicate of 500. The percent identity of the plant reductases and dehydrogenases used in the tree were calculated using the Needleman-Wunsch Global Alignment tool from NCBI with the gap costs set at an existence of 11 and an extension of 2.