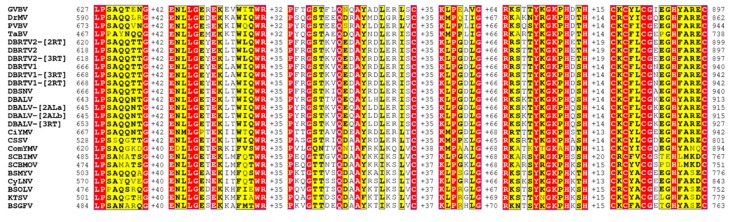
Figure 5.
Comparison of highly conserved amino acid residues in the coat protein encoded by the N-terminal half of the ORF 3 product among DBALV-[2ALa], DBALV-[2ALb], DBRTV1, DBRTV1-[2RT], DBRTV1-[2RT], DBRTV2, DBRTV2-[2RT], DBRTV2-[3RT], DBALV-[3RT], DBALV, DBSNV, 14 badnavirus and other members of the family Caulimoviridae (see text for detail). Sequences were aligned with CLUSTAL OMEGA (http://www.ebi.ac.uk/Tools/msa/clustalo/) and coloured using ESPript 3.0 [57], where functional conserved amino acid residues are highlighted with yellow backgrounds and complete consistent residues with red backgrounds. Numbers of the starting and ending amino acids are specified before and after each sequence, respectively. The numbers of residues (gaps) between blocks are presented.