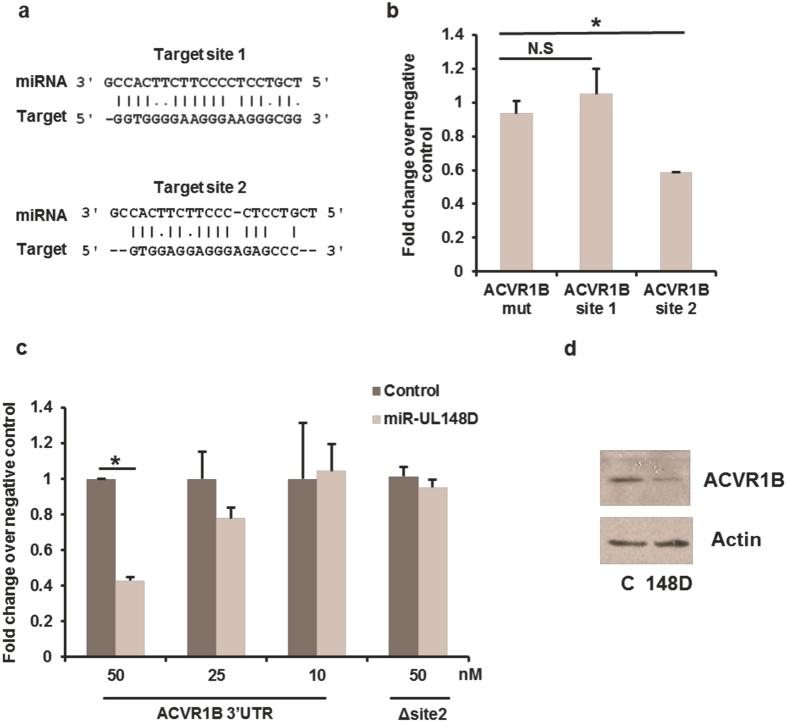
Figure 3. ACVR1B 3′ UTR is directly repressed by miR-UL148D at one of two predicted target sites.
The computer algorithms Reptar and RNAhybrid predict miR-UL148D to target the ACVR1B 3′ UTR at two separate non-canonical sites; both are shown in (a). (b) 293T cells were transfected with a negative control siRNA or miR-UL148D mimic together with a construct containing either an irrelevant sequence (ACVR1B mut), ACVR1B target site 1 or 2 was inserted in the 3′ UTR of the luciferase reporter gene. At 24 hours post transfection, luciferase activity was determined, and fold change in luciferase activity in the miR-UL148D transfected cell compared to negative control is displayed. (c) 293T cells were transfected with a negative control siRNA or miR-UL148D mimic at concentrations indicated together with a luciferase reporter vector containing the entire ACVR1B 3′UTR with (ACVR1B 3′UTR) or without (Δ site 2). Luciferase activity was determined at 24 hours post transfection and data is shown as described in (b). Significant differences were determined using t-test and marked by (*p < 0.05) whilst N.S. denotes not significant differences. (d) KG-1 cells were transfected with negative control siRNA (C) or miR-UL148D mimic (148D), after which ACVR1B levels were assessed by western blot at 48 hours post transfection. The autoradiograph shown has been cropped with no further manipulation.