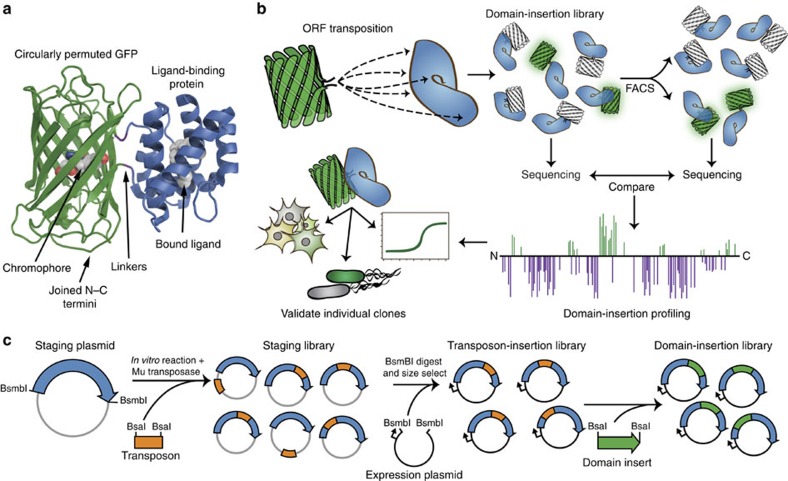
Figure 1. High-throughput biosensor construction using DIP-seq.
(a) Illustration of a single-FP metabolite protein biosensor. cpGFP is fused to a LBD in a manner such that metabolite binding by the LBD causes a change in fluorescence of the attached cpGFP. (b) Overview of the domain-insertion profiling method used to create and identify functional biosensors. A diverse library of fusions, with cpGFP inserted into a LBD, is created and screened with FACS. Initial and sorted libraries undergo NGS analysis and these data are used to identify insertion sites within the LBD that are enriched during screening. Clones of interest are individually tested to validate biosensor functionality. (c) Method of domain-insertion library creation. An engineered transposon containing a selectable marker is inserted into a staging plasmid carrying the LBD ORF using an in vitro transposase reaction. Staging plasmids with an insertion are selected for, purified and digested with an enzyme that releases the LBD ORF from the staging backbone (grey). LBD ORFs with an inserted transposon are size-selected and cloned into an expression plasmid (black). Finally, a domain of interest (in this paper, cpGFP) is inserted into the cloning site created by the modified transposon.