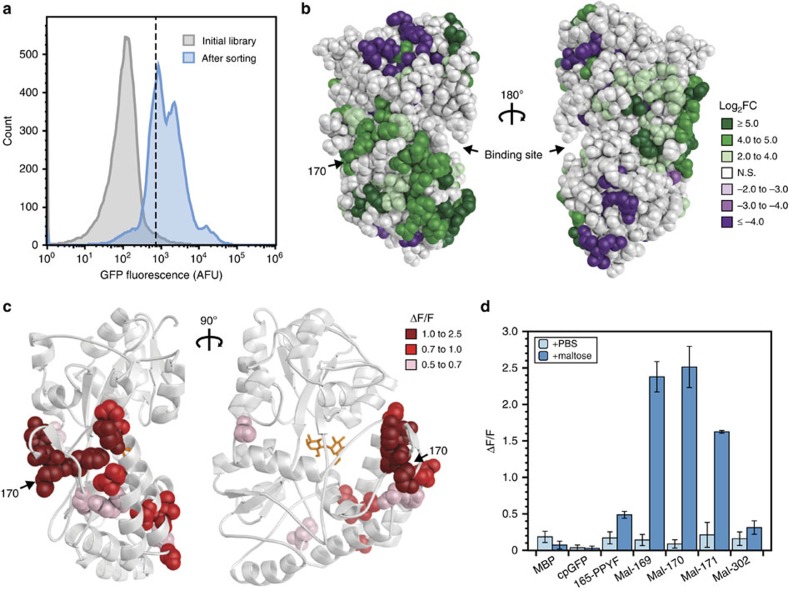
Figure 2. Maltose-binding protein as proof-of-concept target of random domain insertion.
(a) Fluorescence histograms of representative MBP-cpGFP libraries. Induced libraries were sorted via FACS with gating set to collect cells above a threshold GFP fluorescence level shown with a dashed line. (b) Enrichment of cpGFP-insertion sites (log2 of fold change) from FACS mapped onto MBP crystal structure (PDB 1ANF). Enrichment calculated from NGS, comparing pre- and post-sort libraries. Insertion sites that went from undetectable to detectable were set at +6, while sites that were cleared from the library (not sequenced in post-sort library) were set at −6. Amino acid 170 is indicated with an arrow. (c) Functional biosensor insertion sites mapped onto the MBP structure. Sites highlighted with sphere representation of side chains, with colours showing level of activity (ΔF/F=(Fligand−F0)/F0). (d) Activity assay of individual constructs. Fluorescence change on addition of saturating maltose (1 mM) or PBS was measured, shown by mean±s.d. of ΔF/F (three biological replicates). Sample 165-PPYF is the previously published construct EcMBP165-cpGFP.PPYF.T203V (ref. 19).