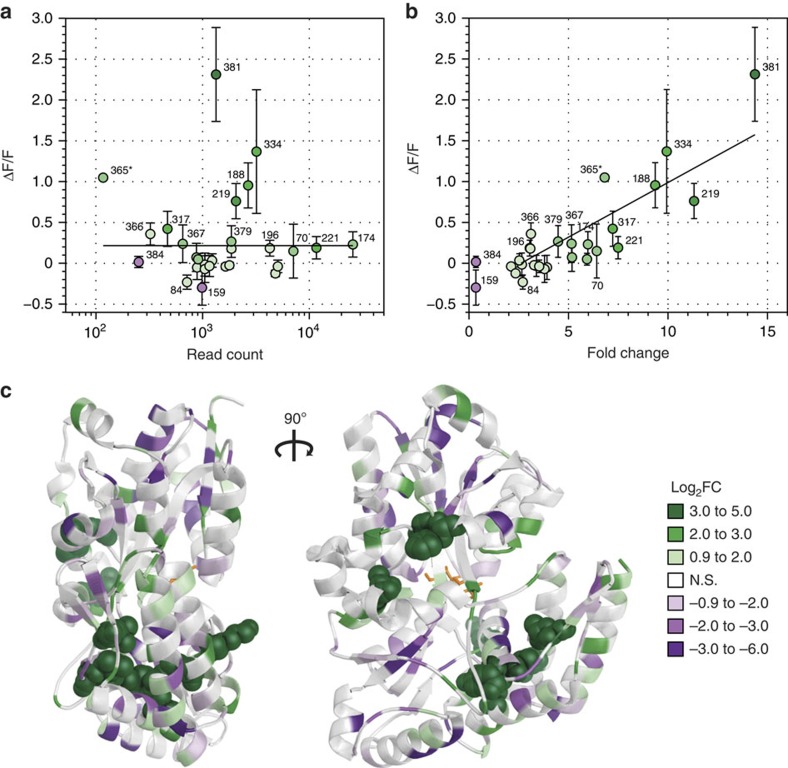
Figure 4. DIP-seq with successive rounds of positive and negative selection enriches for functional biosensors.
(a) Activity versus final NGS read count. Activity is reported as mean±s.d. of ΔF/F from at least three biological replicates (asterisk denotes one sample with two replicates). Read count is combined from two biological replicates from the final sort 3 library. Activity is not correlated with final read count (Pearson's R2=8.6 × 10−5, P=0.96). (b) Activity versus enrichment. Activity is as shown in a. Enrichment, shown as fold change, is calculated from two biological replicates comparing the initial and final library NGS read counts. Calculated fold change with P<0.1 shown. Activity is linearly correlated with enrichment (Pearson's R2=0.73, P=5.3 × 10−11). Colours of points in a,b are binned based on fold-change values after all sorting rounds. (c) Enrichment of cpGFP-insertion sites (log2 of fold change) mapped onto TMBP crystal structure (PDB 1EU8). Sites cleared from the library are set at a value of −5. Colours are binned for enrichment values, with white representing sites of nonsignificant enrichment (P⩾0.1). Sites in the top bin, having the highest enrichment, are highlighted by sphere representation of their side chains.