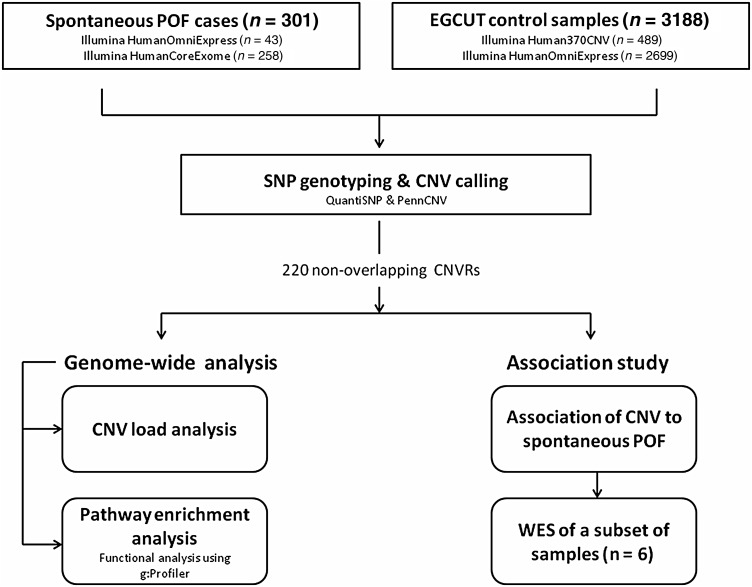
Figure 1.
Schematic representation of study design. A total of 301 spontaneous premature ovarian failure (POF) patients were enrolled in the study. After single nucleotide polymorphism (SNP) genotyping and copy number variation (CNV) calling, genome-wide CNV load analysis was performed per individual to estimate the effect of CNV load in the genome on time of menopause. Subsequently, all the detected CNVs were compared against the EGCUT control population (n = 3188) to identify nonoverlapping CNV regions (CNVRs) (n = 220), for which functional enrichment and association studies were performed. After the following interpretation of each CNVR, six individuals with hemizygous deletions were also selected for whole-exome sequencing (WES) to determine whether identified deletions can result in unmasking recessive mutations or other POF-associated variants in the exome.