Figure 1.
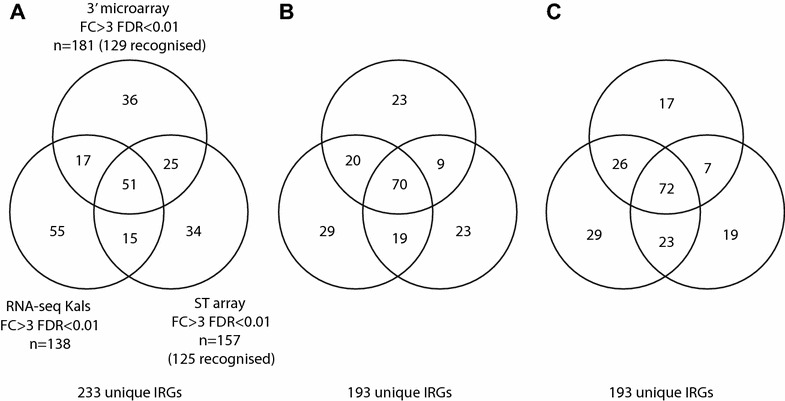
Correlation of ISGs identified as significant by RNA-seq and microarray. Venn diagrams showing correlation of significant ISGs (FC ≥ 3; FDR ≤ 0.01, unless stated otherwise) for: (i) Illumina 100b paired-end RNA-seq, (ii) Affymetrix 32K GeneChip Chicken Genome Array and (iii) Chicken Gene 1.0 ST Array, following induction of CEF for 6 h with 1000 units of rChIFN1. (A) Total hits (“n =”) shown for each technology; those corresponding to Galgal4 assembly Gene IDs are shown in brackets (“recognised”)—RNA-seq hits all represent Galgal4 mapped genes. (B) Hits from array technologies were manually curated to maximise numbers of corresponding genes. (C) Curated array hits shown in (B) that are present amongst RNA-seq hits, but at lower levels of significance, were transferred to the respective RNA-seq-overlapping sectors. Total genes are shown for (A–C).
