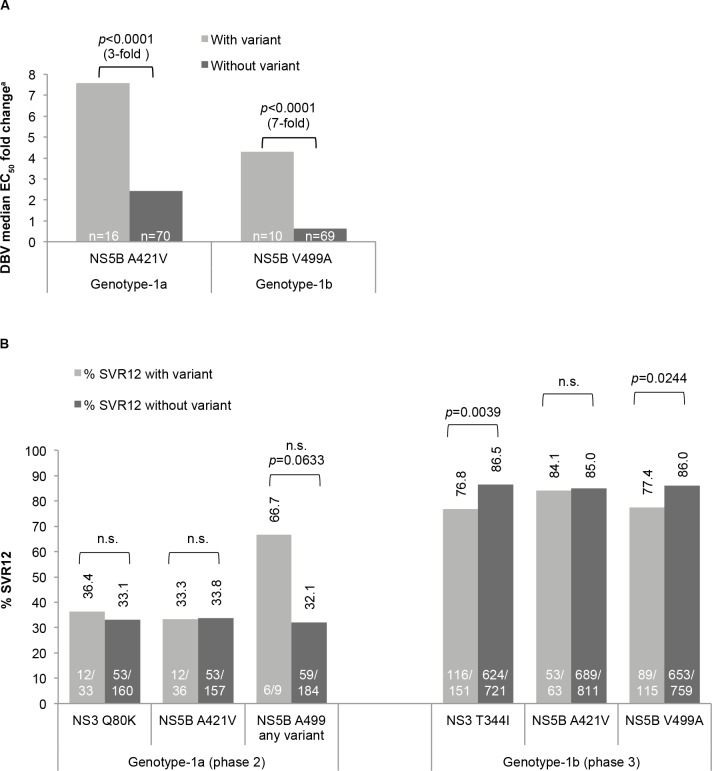
Fig 1.
Impact of baseline HCV polymorphisms on (A) in vitro deleobuvir susceptibility and on (B) response to treatment with faldaprevir/deleobuvir/RBV. Data pooled from (A) 1241.2, 1241.7, SOUND-C1, and SOUND-C2; and (B) SOUND-C2 (RBV-containing arms only), SOUND-C3 (phase 2 data), and HCVerso1 and 2 (phase 3 data). Statistical analyses are described in ‘Materials and Methods’. GT-1a ‘NS5B A499 any variant’ includes A499T (n = 4), A499A/S, and A499V. aFold change with respect to subtype reference EC50 of sub-genomic replicons (GT-1a = 17.5 nM [n = 19] and GT-1b = 11.5 nM [n = 49]). DBV, deleobuvir; EC50, 50% effective concentration; HCV, hepatitis C virus; n.s., not significant (p>0.05); RBV, ribavirin; SVR12, sustained virologic response 12 weeks after treatment.