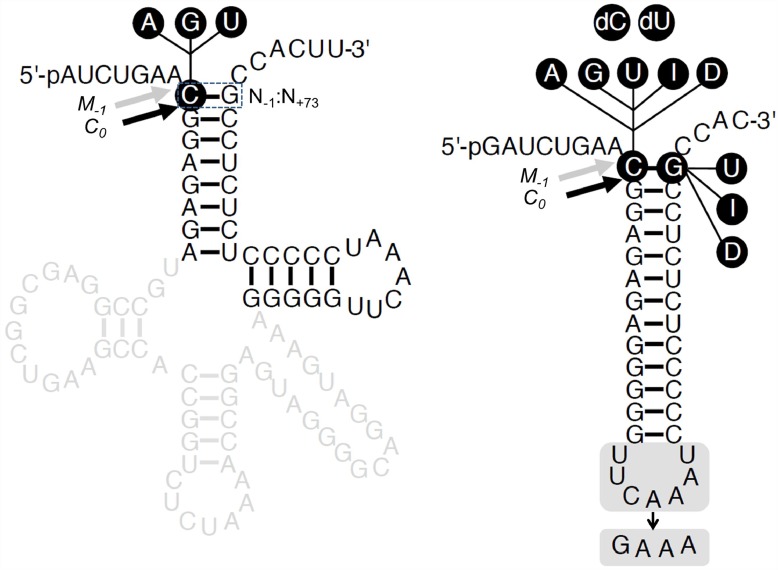
Fig 1. Secondary structures of substrates used in this study.
Secondary structures of pSu1 and pATSer. The highlighted regions/residues were substituted to generate the different variants as indicated, A, adenosine, G, guanosine, U, uridine, I (Ino), inosine and D (DAP), 2,6-diaminopurine; dC, deoxycytosine; and dU, deoxyuridine. The canonical RNase P cleavage sites between residues N-1 and N+1 (correct cleavage denoted C0), and the alternative cleavage sites between residues N-2 and N-1 (miscleavage denoted as M-1) are marked with black and grey arrows, respectively. The N+73 position, which immediately precedes the 3'-terminal CCA-motif, corresponds to the discriminator base.