Abstract
Introduction
Bronchiolitis is a major public health problem worldwide. However, no effective treatment strategies are available, other than supportive care.
Areas Covered
Although bronchiolitis has been considered a single disease diagnosed based on clinical characteristics, emerging evidence supports both clinical and pathobiological heterogeneity. The characterization of this heterogeneity supports the concept that bronchiolitis consists of multiple phenotypes or consistent grouping of characteristics.
Expert Commentary
Using unbiased statistical approaches, multidimentional clinical characteristics will derive bronchiolitis phenotypes. Furthermore, molecular and systems biology approaches will, by linking pathobiology to phenotype, identify endotypes. Large cohort studies of bronchiolitis with comprehensive clinical characterization and system-wide profiling of the “-omics” data (e.g., host genome, transcriptome, epigenome, viral genome, microbiome, metabolome) should enhance our ability to molecularly understand these phenotypes and lead to more targeted and personalized approaches to bronchiolitis treatment.
Keywords: bronchiolitis, lower respiratory infection, infants, phenotypes, endotypes, immune response, genome, transcriptome, microbiome, systems biology
1. INTRODUCTION
Bronchiolitis is the most common lower respiratory infection in young children [1]. Although “bronchiolitis” refers to inflammation of the bronchioles, this inflammation is inferred in young children who have respiratory distress with signs of an acute, viral, lower respiratory infection. Consequently, bronchiolitis remains a clinical diagnosis without an international consensus [2–4]. In the 2014 American Academy of Pediatrics (AAP) guidelines, bronchiolitis was defined as a constellation of clinical signs and symptoms occuring in children yougner than 2 years of age, including a viral upper respiratory tract prodrome followed by increased respiratory effort and wheezing [2]. By contrast, most non-U.S. clinicians and bronchiolitis researchers believe that the AAP definition is too broad because the distinction between bronchiolitis and viral-induced recurrent wheeze becomes increasingly difficult, if not impossible, as the child ages [1].
The classification system of human diseases – i.e. the “diagnostic labels” we also apply to bronchiolitis – was established by Sir William Osler in the 19th century, on the basis of the principal organ in which signs and symptoms manifest with some anatomical, physiological, and pathological correlates [5]. However, there is a growing concern that this “Oslerian paradigm” may overgeneralize disease phenotypes and cannot individualize diagnosis or management according to their molecular pathway [5,6]. Current diagnosis and management guidelines of bronchiolitis [2–4] are based on this paradigm and do not consider novel molecular information, which are now available with the advent of technology.
A growing body of evidence, including our own research, now suggests that “bronchiolitis” likely represents a continuum of different diseases that may share biological mechanisms (endotypes) and present with similar clinical features (phenotypes) that may require individualized treatment. To develop phenotype-specific treatment strategies, integration of clinical and molecular information (e.g., genomic information) is needed to achieve comprehensive understanding of the pathobiological mechanisms that underlie bronchiolitis phenotypes. The objective of this review is to provide an overview of the evidence supporting the heterogeneity of bronchiolitis, to highlight recent insights into different mechanisms of bronchiolitis pathogenesis, and to allude to systems biology approaches, some borrowed from other fields (e.g., childhood asthma), that might enable identification of bronchiolitis endotypes.
2. EPIDEMIOLOGY OF BRONCHIOLITIS
Bronchiolitis is a major public health problem in the U.S. and worldwide [7–11]. Almost all children are exposed to respiratory syncytial virus (RSV) and other causative pathogens of bronchiolitis (e.g., rhinovirus) during the first two years of life [2]. Of these, approximately 40% of children develop clinical bronchiolitis [12,13]. Most children with bronchiolitis have mild illness; however, some children present to the emergency department (ED), and others require hospitalization (severe bronchiolitis) [1]. In the U.S., bronchiolitis is the second leading cause of ED visits among infants, accounting for 15% of all infant ED visits [8,10]. Furthermore, bronchiolitis is the leading cause of hospitalizations, accounting for 18% of all infant hospitalizations (approximately 130,000 hospitalizations annually), with a direct cost of $550 million annually [9]. Severe bronchiolitis has a peak incidence between two and six months of age. Risk factors for severe bronchiolitis include younger age (<12 weeks), prematurity, environmental factors (such as passive smoking, crowded household, high altitude), and comorbidities (such as congenital defects of the airways, chronic lung disease, congenital heart disease, immunodeficiency, neurologic disease) [1].
3. HETEROGENEITY OF BRONCHIOLITIS
Despite the large public health burden, no effective treatment strategies are available, other than supportive care, in young children with bronchiolitis [2–4]. Current evidence is largely based on clinical trials of specific interventions (e.g., bronchodilators, corticosteroids) with mixed results, and an underlying assumption that bronchiolitis is a single disease entity with similar clinical characteristics, causes, and mechanisms [1]. As the inferences were based on the group mean data from populations that might include different subgroups [14,15], even after excluding large numbers of children with physician-diagnosed bronchiolitis (e.g., children with non-RSV bronchiolitis, history of prior breathing problems, age older than 1 year), the current evidence might have failed to encompass differences in efficacy and safety profiles.
Indeed, there is emerging evidence to suggest that bronchiolitis is not homogeneous. For example, most children with bronchiolitis have a mild-to-moderate disease course while approximately one in ten of these children require hospitalization [1] and 2% of whom undergo mechanical ventilation [9]. Furthermore, there is a wide range of time to recovery [16] and variable risk of relapse [17]. Epidemiologic research also has documented that the susceptibility and severity vary widely by clinical characteristic, such as chronologic age, racial/ethnicity, and coexisting conditions (e.g., prematurity, chronic lung disease of prematurity, congenital heat disease, immunodeficiency, and neurologic disease) [1]. Additionally, the disease severity also varies by environmental factors, such as nutrition and passive smoking [1,18]. Furthermore, several studies have documented a diverse group of respiratory viruses involved in bronchiolitis, as a sole pathogen or co-infecting pathogen, and their contribution to varied clinical outcomes [1,19,20] – e.g., associations between sole rhinovirus infection and a shorter hospital length-of-stay [19,21,22] and between RSV/rhinovirus coinfection and a longer hospital length-of-stay [19] when compared to sole RSV infection. Viral load adds further complexity; studies have demonstrated a wide range of RSV viral load and its relation with bronchiolitis severity [23]. Finally, studies have reported a variability in treatment response among subgroups [24]. For example, although a meta-analysis of systemic corticosteroids based on the group mean data from populations with bronchiolitis did not show superior efficacy over placebo [15], a recent trial of infants with eczema or a family history of asthma in a first-degree relative demonstrated that dexamethasone reduces the time to readiness for discharge in severe bronchiolitis [24].
These studies collectively indicate that bronchiolitis is a heterogeneous condition. The term bronchiolitis, similar to “asthma” [25], is an umbrella-like term that equates to a definition of grouped host, microbial, clinical, and physiological characteristics (Figure 1). In poorly understood ways, grouped features contribute to severity, which also becomes part of the definition. We believe that these characteristics can identify multiple subgroups or phenotypes – the observable properties of an individual that are produced by the interactions of the host genetic predisposition and the environment, and that identifying these phenotypes is of critical importance in order to develop effective treatment and preventive interventions [25].
Figure 1. Schematic representation of the umbrella term “bronchiolitis”.
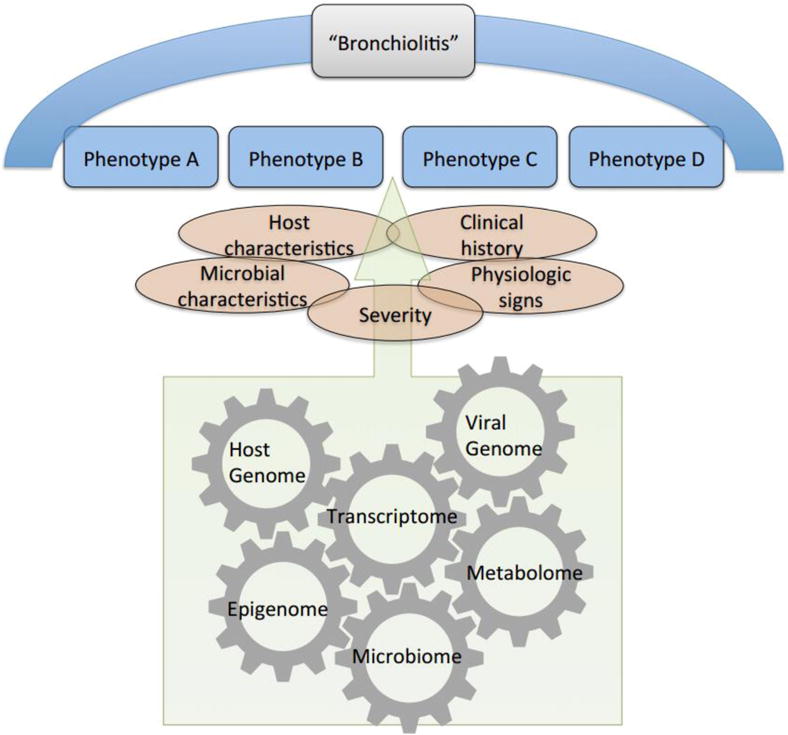
The term “bronchiolitis” likely represents a continuum of different diseases that may present with similar clinical features (phenotypes) and share biological mechanisms (endotypes) involving host genome, transcriptome, epigenome, virus genome, microbiome, and metabolome. These phenotypes and endotypes remain under investigation.
4. PHENOTYPING BRONCHIOLITIS
Previous studies examining subgroups of children with bronchiolitis generally classify individuals according to a single dimension (e.g., RSV status) or to a limited number of characteristics (e.g., RSV status and history of prior breathing problems) – a “biased” approach. However, given the complex, high-dimensional interactions between host, environment, and microbial factors in bronchiolitis, we believe that “unbiased” (or “hypothesis-free”) statistical approaches would be useful to address disease heterogeneity, through identifying more homogeneous subgroups of children sharing similar characteristics of the disease. Compared to the biased approaches, the benefits of unbiased multidimensional approaches in addressing different phenotypes have been demonstrated for other respiratory disorders, such as childhood asthma [25–28]. An example of the unbiased statistical approach is latent class analysis. This method statistically identifies distinct groups of subjects (latent classes) sharing similar characteristics, although the clinical features that go into the model are biased by a priori knowledge of the disease. To the best of our knowledge, there are no published research using unbiased approaches to children with bronchiolitis other than preliminary data from a multicenter cohort of 2,207 U.S. children with severe bronchiolitis which demonstrated four distinct clinical profiles [29]. Profile A was characterized by history of wheezing and eczema, wheezing at the ED presentation and rhinovirus infection. Profile B included children with wheezing at the ED presentation, but, in contrast to profile A, most did not have history of wheezing or eczema; this profile had the largest probability of RSV-infection. Profile C was the most severely ill group, with longer hospital stay and moderate-to-severe retractions. Profile D had the least severe illness, including non-wheezing children with shorter length-of-stay. We recently validated these results in a similar, but entirely separate, cohort of 408 Finnish children with severe bronchiolitis (unpublished data). The concept of bronchiolitis heterogeneity is now evolving from one that focuses on a limited number of clinical characteristics (e.g., RSV bronchiolitis) to one that phenotypes bronchiolitis using a comprehensive multidimensional approach.
5. ENDOTYPING BRONCHIOLITIS
To develop phenotype-specific treatment strategies, the concept of phenotype should be further evolved to that of the “endotype” – i.e., specific pathobiological mechanisms that underlie the observable properties of a phenotype [25]. This approach is being applied by many different groups to approach other airway inflammatory diseases, such as asthma [30–35]. The identification of endotypes necessitates an integration of phenotypic information and molecular data, which the increasing availability of high-throughput “-omics” technologies has enabled us to characterize – e.g., host genome, transcriptome, epigenome, viral genome, microbiome, and metabolome (Table 1).
Table 1.
Glossary
| 16S rRNA gene sequence | Sequencing DNA within the hyper-variable regions of the 16S ribosomal RNA (16s rRNA) gene that enables identification of bacteria and archaea. |
| Genome | The complete set of genomic information for an organism including genes and non-coding sequences. |
| Epigenome | The collection of DNA modifications that affect gene expression and occur without direct alteration of the DNA sequence (e.g., DNA methylation, histone modification, and microRNAs). |
| Microbiome | The collection of commensal, symbiotic, and pathogenic microbes (e.g., bacteria, archaea, fungi, viruses) and their genomes in the human body. |
| Microbiota | All microbes that are found in a particular niche or region. |
| Metabolomics | High-throughput characterization of metabolites in body fluids (e.g., plasma, serum, exhaled breath condensate, and urine). |
| Metagenomic sequencing | Sequencing the total DNA of the ecosystem, with the advantage of providing information on the presence of bacteria, archaea, DNA viruses, eukarya, and their functionality. |
| Transcriptome | The complete set of RNA molecules produced in one cell or a population of cells. |
| Systems biology | An approach, by modeling diverse types of high-dimensional interactions, to developing a more comprehensive understanding of biology at multiple scales (molecular, cellular, tissue, organ, organism, and community). |
5.1. Host Genome
Compared to other airway diseases, there are very few genetic studies of bronchiolitis. However, the limited literature suggests a relationship of host genetics with susceptibility and severity of bronchiolitis [36]. For example, a Danish twin study documented that genetic factors account for 16% of the individual susceptibility to develop severe RSV bronchiolitis [37]. Although no genome-wide association study (GWAS) has examined bronchiolitis, candidate gene association studies have provided evidence for genes that increase the severity of RSV infection along many biological pathways, such as innate immunity, adaptive immunity, chemotaxis, airway epithelial response, and known allergic asthma genes [36,38,39]. For example, Janssen et al., by examining 384 single-nucleotide polymorphisms from 220 genes involved in immune responses, found that genes from innate immune pathway are important in RSV infection severity [40]. Additionally, Wu et al., by examining 374 term infants enrolled in the Tennessee Children’s Respiratory Initiative (TCRI) study, found an association of β2-adrenergic receptor gene polymorphisms with bronchiolitis severity score in African American infants [41]. These studies not only provide a potential mechanism to explain the variable response to bronchodilator in children with bronchiolitis but also indicate the multiplicity of underlying mechanisms, such as innate and adaptive immunity, that might explain the heterogeneity of the disease.
5.2. Transcriptome
Transcriptomics is the systematic and unbiased characterization of RNA transcripts across the genome [42]. A disease-relevant tissue (e.g., airway epithelium) is sampled, and then oligonucleotide microarrays or RNA sequencing are used to profile the tissue-specific RNA transcripts. Transcriptomics offers a complementary approach to genome studies to examine diseases because RNA transcripts present the more dynamic process in the tissue involved in the disease pathogenesis.
Several studies have applied these transcriptome approaches to children with bronchiolitis. By investigating whole blood [43–46] and upper airway gene expression profiles [47], researchers demonstrated overexpression of interferon-related pathway in children with severe RSV infection compared to healthy controls or mild RSV infection. Furthermore, Mejias et al., by examining whole blood transcriptome from children with severe bronchiolitis either by RSV, rhinovirus, or influenza in the U.S. and Finnish cohorts, observed pathogen-specific transcriptional signatures [43]. Specifically, children with RSV infection had overexpression of neutrophil-related genes and suppression of B cell, T cell, lymphoid lineage, and antimicrobial response genes while those with rhinovirus infection had a higher expression of cytotoxic/natural killer (NK) cell genes. These transcriptomic findings suggest that mechanisms underlying bronchiolitis pathogenesis differ, at least, by causative virus. Additionally, compared to mild-to-moderate infection, severe RSV infection was associated with an overexpression of neutrophil and inflammation genes as well as an under-expression of T cell, cytotoxic and plasma cell genes, indicating the important contribution of host immune response to the clinical course [43].
5.3. Epigenome
Alteration in gene expression can occur without direct change in the DNA sequence. The study of such epigenetic change (e.g., DNA methylation, histone modification, and microRNAs [miRNAs]) characterizes DNA sequence-independent modification that contributes to transcriptomic variations and downstream phenotypes [42]. In particular, miRNAs – a class of noncoding single-stranded RNA molecules approximately 18–25 nucleotides in length – have emerged as important gene expression regulators. By affecting post-transcriptional expression, miRNAs regulate the expression of at least 30% of human protein-encoding genes and play a modulatory role in host immune response [48]. Indeed, emerging evidence has shown that miRNAs participate in the maintenance of the airway epithelial barrier and are also implicated in the modulation of antiviral defense in epithelial cells. For example, Leahy et al., by investigating miRNA expressions in NK cells in children with severe bronchiolitis and healthy controls, reported that miRNA profile is distinct in bronchiolitis and that the targets of deregulated miRNAs include NFκB on the IL-15 signaling pathway [49]. Additionally, studies using experimental models have demonstrated that respiratory viruses (e.g., RSV, rhinovirus, and influenza virus) induce distinct miRNA profiles that have different target genes and functions [48,50–54]. These data suggest that epigenetic regulation pathways of host defense against respiratory infections differ by causative virus.
5.4. Viral Genotype
As with host genetic variations, viral genetic differences play a role in disease pathogenesis. Across virus families, studies have reported that relatively minor changes in viral genome have a large impact on pathogenesis – for example, the virulence of the 1918 influenza hinges on a few amino acids [55]. Similarly, emerging evidence indicates that genotypes of RSV and rhinovirus – the two most common causative viruses of bronchiolitis [1] – have an impact on both the pathogenesis and severity of bronchiolitis [56].
RSV, a member of the Paramyxoviridae family of RNA viruses, has one serotype, within which there are two antigenic subgroups, A and B. Within these subgroups, RSV strains can be further classified into clades (GA1-GA7, GB1-GB4) based on the sequence of a hypervariable region of the G gene [56]. Studies reported that antigenic subgroup A, especially clade GA3, is associated with a greater clinical severity of RSV infection [57,58]. In experimental models using well-differentiated pediatric bronchial cells, GA5 isolate causes high degree of epithelial sloughing and goblet cell hyperplasia [59]. In addition, the molecular determinants of RSV strain-specific virulence are an active field of research. Several studies suggested that strain variations in the RSV fusion (F) protein sequence lead to a range of fusion activity, cytopathology, neutrophil responses, TH2 immune responses, and epithelial injury [56].
Rhinoviruses, members of the Picornaviridae family, have long been thought as the major cause of upper respiratory infection. However, recent studies have demonstrated that rhinoviruses also cause asymptomatic infections and lower respiratory infections, including bronchiolitis in children [1,60,61]. Rhinoviruses are highly genetically and antigenically diverse. Indeed, among the three species groups (A, B, and C), sequencing the rhinovirus capsid genes revealed >150 types [56]. Although some studies reported no significant between-species difference in bronchiolitis severity [61], emerging evidence suggests that the rhinovirus C species are most clinically significant. For example, the rhinovirus C species are the most common rhinovirus group associated with lower respiratory infections [62–64] in the ED and inpatient settings. Additionally, Lee et al. reported, in the Childhood Origins of ASThma (COAST) cohort in the U.S., that rhinovirus A and C species are more likely to cause severe respiratory infection in infants, compared to rhinovirus B species, in the outpatient setting [65]. This study also reported a within-species difference in virulence – e.g., some C types being more virulent than the other C types, suggesting that rhinovirus genotype-specific pathogenesis extends beyond the species-level to the type-level [65].
5.5. Microbiome
Although bronchiolitis is typically caused by a viral infection, emerging evidence indicates that microbes inhabiting the human body (the microbiota) play an important role in bronchiolitis pathogenesis. Over the past decade, the use of 16S rRNA gene and metagenomic sequencing of aggregate microbial genomes (the microbiome) made it possible to profile the collective microbial community, and revealed more microbiota and phylogenetic relationships than previously detectable [66].
Although few studies have specifically investigated infants with bronchiolitis [67–69], emerging evidence shows that airway microbiome may influence immune responses in the airway [66,70–74], suggesting a role of airway microbiome in the development and morbidity of acute respiratory infections [75–80]. For example, Kloepfer et al., by applying quantitative polymerase chain reaction technique to nasal samples of 308 U.S. children in the RhinoGen cohort, found that Moraxella catarrhalis and Streptococcus pneumoniae together with rhinovirus infection contributes to a higher severity [77]. By applying a culture-dependent technique to 265 infants from the Copenhagen Prospective Studies on Asthma in Childhood2000 (COPSAC2000) cohort, Vissing et al. found that 1-month-old infants with colonization of M. catarrhalis or Haemophilus influenzae in hypopharynx had an increased risk of subsequent development of bronchiolitis [79]. Similarly, Teo et al., by using 16S rRNA gene sequencing in 234 Australian infants from the Childhood Asthma Study cohort, demonstrated that Haemophilus-dominant nasopharyngeal microbiota was associated with a higher incidence of acute respiratory infection and higher severity in infants with high risk of atopy [75].
Recently, we applied 16S rRNA gene sequencing technique to nasopharyngeal samples in the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure (INSPIRE) study, a population-based birth cohort of 1,952 healthy infants, and found marked differences in microbiota diversity and richness between healthy infants and those with acute RSV infection. For instance, the more abundant genera during acute RSV infection were Corynebacterium, Haemophilus, Moraxella, and Streptococcus [81]. Additionally, in the 35th Multicenter Airway Research Collaboration (MARC-35) study, a multicenter cohort of approximately 1,000 U.S. infants hospitalized for bronchiolitis, we found viral-microbial relationships in the airway. For example, infants with RSV infection had a high abundance of Streptococcus and a low abundance of Haemophilus and Moraxella genera while infants with rhinovirus infection had the opposite pattern [68]. Furthermore, in unpublished work, we applied an unbiased clustering approach and identified four distinct microbiota profiles – three profiles were dominated by either Haemophilus, Moraxella, or Streptococcus genus, while the fourth profile had highest bacterial richness. The rate of intensive care use was highest in infants with Haemophilus-dominant profile and lowest in those with Moraxella-dominant profile. These findings were externally validated in a similar, but entirely separate, cohort of 307 U.S. children hospitalized for bronchiolitis (unpublished data).
Although these findings suggest a complex interplay between viral pathogen, airway microbiome, and bronchiolitis pathogenesis, the nature of this microbial association requires clarification. It is possible that there is a causal relationship – i.e., a specific microbiome profile (e.g., Haemophilus-dominant microbiome) in the infant’s airway alters immune responses to increase the severity of bronchiolitis. Alternatively, Haemophilus-dominant microbiome may be simply a marker of an infant who is prone to develop more-severe bronchiolitis. Additionally, reverse causation – i.e., more-severe illness results in a rapid overgrowth of specific bacteria– is also possible. These possibilities are not mutually exclusive. Notwithstanding the complexity, these data indicate the heterogeneity of airway microbiome profiles and its potential contribution to the pathobiology of bronchiolitis.
5.6. Metabolome
Metabolomics systematically identify and quantify the collection of metabolites in biological specimens by using nuclear magnetic resonance or liquid chromatography mass spectrometry. Metabolomics profiling provides a snapshot of dynamic physiology and has been used as a tool of biomarker discovery [42]. To date, no study has applied a metabolomic approach specifically to children with bronchiolitis. However, within the limited literature about metabolome in infants, Herberth et al. performed a metabolome analysis in sera of 495 healthy newborns in Germany, and found a relationship between high hexose levels, increased expression of NLRP3 inflammasome and effector cytokine IL-1β, and an increase risk of parent-reported wheezing illness by age 2 years [82]. In other disease conditions, such as asthma, metabolome studies using exhaled breath condensate, serum, and urine specimens have identified phenotype-specific metabolic profiles, such as eosinophilic and neutrophilic asthma [42].
6. SYSTEMS BIOLOGY APPROACH
Data generated through host genome, transcriptome, epigenome, viral genome, microbiome, and metabolome studies have advanced our understanding of the various mechanisms that underlie bronchiolitis pathogenesis. However, as a heterogeneous disease, it is unlikely that a single molecular profiling modality can capture the interdependent dynamics of the molecular networks involved in bronchiolitis (Figure 1). We believe that integrating these multiscale data with phenotypic information is a necessary step to better understand bronchiolitis. Systems biology approach is attractive as it has the potential to model the multidimensional interactions between these factors that ultimately lead to diverse disease phenotypes and treatment responses across individuals [42].
Although no study to date has utilized a systems biology approach to identify bronchiolitis endotypes, a few studies have applied this approach to childhood asthma [30,31]. For instance, by using multi-step decision tree method to integrate clinical, physiological, immunological, and transcriptomic data, George et al. identified several endotypes (e.g., atopic, mixed eosinophilic and neutrophilic, and TH2-low/metabolic syndrome-related endotypes) in 192 U.S. children with asthma [30]. In addition, probabilistic causal network method, another systems biology approach, has been applied to several other diseases, such as obesity [42] and provided mechanistic understanding. This approach infers causal relationships between molecular interactions by constructing a consensus model that best fits the data and identifies directionality of relationships between the tens of thousands of molecular variables [83]. The implementation of such systems biology approaches in bronchiolitis is our next challenge.
The success of “-omics” and systems biology approaches depends on our ability to manage and interpret large-scale multi-dimensional data. To obtain meaningful results, processing and integrating high-dimensional data require multiple steps, such as data transfer and management, access control, data format standardization, and appropriate modeling of biological systems via data integration. Robust bioinformatics and computational infrastructures are also warranted for each of these steps. Additionally, other potential problems, such as bias (selection, measurement, confounding) and reproducibility issue, should be minimized by careful control, planning of the experiments, and validation, as is typically done in high-quality epidemiological research.
7. EXPERT COMMENTARY
With the advent of high-throughput molecular techniques, our understanding of bronchiolitis pathogenesis has greatly improved. A growing number of studies challenge the conventional wisdom that bronchiolitis is a homogeneous condition and the traditional term “bronchiolitis” appears increasingly out-of-date. With continued advances, it may become possible to create more detailed clinically and molecularly focused definitions of this condition. Using unbiased statistical approaches, multidimentional clinical characteristics will derive bronchiolitis phenotypes. Furthermore, molecular and systems biology approaches will, by linking pathobiology to phenotype, identify endotypes. These effort will not only enhance our ability to molecularly understand these phenotypes but also lead to identification of biomarkers and more personalized approaches to bronchiolitis treatment.
8. FIVE-YEAR VIEW
The use of a multidimensional approach to phenotype children of bronchiolitis has only just started [29]. Furthermore, there is no published study that tries to identify bronchiolitis endotypes by linking multiscale molecular information to phenotypic information. These knowledge gaps provide many opportunities for investigation. In this context, two ongoing, NIH-funded cohort studies – 1) the INSPIRE study, and 2) the MARC-35 study – are collaborating to identify phenotypes and endotypes in infants with bronchiolitis. INSPIRE (U19 AI-095227; Hartert) is a population-based birth cohort of 1,952 healthy infants [84]. MARC-35 (U01 AI-87881; Camargo) is a multicenter prospective cohort of 1,016 infants with severe bronchiolitis that is run by the Emergency Medicine Network (EMNet; www.emnet-usa.org). In both cohorts, investigators collected extensive clinical data and upper airway biospecimens at the acute respiratory illness visits (INSPIRE) and at the bronchiolitis hospitalization (MARC-35). Collaboration of the INSPIRE and MARC-35 cohorts has created the one of the largest bronchiolitis consortiums in the world, with a full spectrum of acute severity – from a population-based cohort of infants with bronchiolitis that only required outpatient visit to hundreds of infants with bronchiolitis requiring mechanical ventilation. Importantly, the INSPIRE cohort also includes infants with RSV infection who developed and did not develop bronchiolitis, important comparator groups to understand endotypes of disease and resiliency. Based on the comprehensive clinical and virology data, we will identify acute phenotypes by using a multidimensional approach. Furthermore, we will also define bronchiolitis endotypes through integration of clinical phenotypic and molecular data (e.g., transcriptome, microbiome) by using a systems-biology approach [30,31]. This collaborative effort will not only help us discover new molecular networks involved in bronchiolitis pathogenesis, but also will lead to generation of more refined hypotheses for further investigation. Discovered molecular targets will need to be mechanistically evaluated in experimental model systems (e.g., animal, cell-culture) to further define their functions, associated pathways, and relationship to specific phenotypes. The ultimate test of a phenotype will be investigations of the efficacy of a molecular-targeted intervention [25]. We believe that these combined discovery and hypothesis-driven approaches will support the development of new and effective phenotype/endotype-specific therapies for bronchiolitis.
9. KEY ISSUES.
Despite the large public health burden of bronchiolitis, no effective treatment strategies are available, other than supportive care.
Current evidence on bronchiolitis treatment is largely based on clinical trials with the major assumption that bronchiolitis is a single disease entity with similar disease characteristics and mechanisms.
Recent studies collectively indicate that bronchiolitis is a heterogeneous condition. The term bronchiolitis, similar to “asthma”, equates to a definition of grouped clinical, physiological, and microbial characteristics, which could identify multiple phenotypes.
Previous studies examining subgroups of children with bronchiolitis generally classify individuals according to a single dimension (e.g., RSV status). However, given the high-dimensional interactions between host, environmental, and microbial factors in bronchiolitis, multidimensional statistical approaches would be useful to address disease heterogeneity.
Increasing availability of high-throughput “-omics” technologies has identified multiple mechanisms that underlie bronchiolitis pathogenesis.
Transcriptomic approaches have discovered virus-specific transcriptomic profile in children with bronchiolitis.
Recent microbiome analyses have found heterogeneity of upper airway microbiome profiles, patterns associated with acute viral infection, and its relationship with bronchiolitis susceptibility and severity.
Integrating multiscale “-omics” data with phenotypic information, through a systems biology approach, is a necessary step to identify endotypes and to develop targeted therapies for bronchiolitis.
Acknowledgments
This study was supported by the grants U01 AI-087881, R01 AI-114552, R21 HL-129909, U19 AI-095227, and K24 AI-077930 from the National Institutes of Health (Bethesda, MD). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of Interests
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reference annotations
* Of interest
** Of considerable interest
- 1.Hasegawa K, Mansbach JM, Camargo CA., Jr Infectious pathogens and bronchiolitis outcomes. Exp Rev Anti Infect Ther. 2014;12(7):817–828. doi: 10.1586/14787210.2014.906901. [DOI] [PubMed] [Google Scholar]
- 2.Ralston SL, Lieberthal AS, Meissner HC, et al. Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics. 2014;134(5):e1474–1502. doi: 10.1542/peds.2014-2742. [DOI] [PubMed] [Google Scholar]
- 3.Ricci V, Delgado Nunes V, Murphy MS, Cunningham S, Guideline Development G, Technical T Bronchiolitis in children: summary of NICE guidance. BMJ. 2015;350:h2305. doi: 10.1136/bmj.h2305. [DOI] [PubMed] [Google Scholar]
- 4.Australian Clinical Practice Guidelines. Clinical Guidelines: Management of Bronchiolitis in Children. 2013 [Google Scholar]
- 5.Agusti A, Bel E, Thomas M, et al. Treatable traits: toward precision medicine of chronic airway diseases. Eur Respir J. 2016;47(2):410–419. doi: 10.1183/13993003.01359-2015. [DOI] [PubMed] [Google Scholar]
- 6.Vanfleteren LE, Kocks JW, Stone IS, et al. Moving from the Oslerian paradigm to the post-genomic era: are asthma and COPD outdated terms? Thorax. 2014;69(1):72–79. doi: 10.1136/thoraxjnl-2013-203602. [DOI] [PubMed] [Google Scholar]
- 7.Nair H, Nokes DJ, Gessner BD, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375(9725):1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasegawa K, Tsugawa Y, Brown DF, Mansbach JM, Camargo CA., Jr Temporal trends in emergency department visits for bronchiolitis in the United States, 2006–2010. Pediatr Infect Dis J. 2014;33(1):11–18. doi: 10.1097/INF.0b013e3182a5f324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa K, Tsugawa Y, Brown DF, Mansbach JM, Camargo CA., Jr Trends in bronchiolitis hospitalizations in the United States, 2000–2009. Pediatrics. 2013;132(1):28–36. doi: 10.1542/peds.2012-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasegawa K, Tsugawa Y, Cohen A, Camargo CA., Jr Infectious Disease-related Emergency Department Visits Among Children in the US. Pediatr Infect Dis J. 2015;34(7):681–685. doi: 10.1097/INF.0000000000000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair H, Simoes EA, Rudan I, et al. Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010: a systematic analysis. Lancet. 2013;381(9875):1380–1390. doi: 10.1016/S0140-6736(12)61901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parrott RH, Kim HW, Arrobio JO, et al. Epidemiology of respiratory syncytial virus infection in Washington, D.C. II. Infection and disease with respect to age, immunologic status, race and sex. Am J Epidemiol. 1973;98(4):289–300. doi: 10.1093/oxfordjournals.aje.a121558. [DOI] [PubMed] [Google Scholar]
- 13.Meissner HC. Selected populations at increased risk from respiratory syncytial virus infection. Pediatr Infect Dis J. 2003;22(2 Suppl):S40–44. doi: 10.1097/01.inf.0000053884.21238.13. discussion S44–45. [DOI] [PubMed] [Google Scholar]
- 14.Gadomski AM, Scribani MB. Bronchodilators for bronchiolitis. Cochrane Database Syst Rev. 2014;6:CD001266. doi: 10.1002/14651858.CD001266.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes RM, Bialy LM, Vandermeer B, et al. Glucocorticoids for acute viral bronchiolitis in infants and young children. Cochrane Database Syst Rev. 2013;6:CD004878. doi: 10.1002/14651858.CD004878.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansbach JM, Clark S, Piedra PA, et al. Hospital course and discharge criteria for children hospitalized with bronchiolitis. J Hosp Med. 2015;10(4):205–211. doi: 10.1002/jhm.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa K, Mansbach JM, Teach SJ, et al. Multicenter Study of Viral Etiology and Relapse in Hospitalized Children with Bronchiolitis. Pediatr Infect Dis J. 2014;33(8):809–813. doi: 10.1097/INF.0000000000000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vereen S, Gebretsadik T, Hartert TV, et al. Association between breast-feeding and severity of acute viral respiratory tract infection. Pediatr Infect Dis J. 2014;33(9):986–988. doi: 10.1097/INF.0000000000000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansbach JM, Piedra PA, Teach SJ, et al. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med. 2012;166(8):700–706. doi: 10.1001/archpediatrics.2011.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller EK, Gebretsadik T, Carroll KN, et al. Viral etiologies of infant bronchiolitis, croup and upper respiratory illness during 4 consecutive years. Pediatr Infect Dis J. 2013;32(9):950–955. doi: 10.1097/INF.0b013e31829b7e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jartti T, Aakula M, Mansbach JM, et al. Hospital length-of-stay is associated with rhinovirus etiology of bronchiolitis. Pedatr Infect Dis J. 2014;33(8):829–834. doi: 10.1097/INF.0000000000000313. [DOI] [PubMed] [Google Scholar]
- 22.Marguet C, Lubrano M, Gueudin M, et al. In very young infants severity of acute bronchiolitis depends on carried viruses. PLoS One. 2009;4(2):e4596. doi: 10.1371/journal.pone.0004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasegawa K, Jartti T, Mansbach JM, et al. Respiratory syncytial virus genomic load and disease severity among children hospitalized with bronchiolitis: Multicenter cohort studies in the US and Finland. J Infect Dis. 2015;211(10):1550–1559. doi: 10.1093/infdis/jiu658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alansari K, Sakran M, Davidson BL, Ibrahim K, Alrefai M, Zakaria I. Oral dexamethasone for bronchiolitis: a randomized trial. Pediatrics. 2013;132(4):e810–816. doi: 10.1542/peds.2012-3746. [DOI] [PubMed] [Google Scholar]
- 25.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nature Med. 2012;18(5):716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 26.Fitzpatrick AM, Teague WG, Meyers DA. Heterogeneity of severe asthma in childhood: confirmation by cluster analysis of children in the National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program. J Allergy Clin Immunol. 2011;127(2):382–389. e381–313. doi: 10.1016/j.jaci.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Just J, Gouvis-Echraghi R, Rouve S, Wanin S, Moreau D, Annesi-Maesano I. Two novel, severe asthma phenotypes identified during childhood using a clustering approach. Eur Respir J. 2012;40(1):55–60. doi: 10.1183/09031936.00123411. [DOI] [PubMed] [Google Scholar]
- 28.Garden FL, Simpson JM, Mellis CM, Marks GB, Investigators C Change in the manifestations of asthma and asthma-related traits in childhood: a latent transition analysis. Eur Respir J. 2016;47(2):499–509. doi: 10.1183/13993003.00284-2015. [DOI] [PubMed] [Google Scholar]
- 29.Dumas O, Mansbach J, Hasegawa K, Sullivan A, Piedra P, Camargo CA., Jr Clustering approach to identify bronchiolitis profiles among children hospitalized with bronchiolitis [Abstract] Am J Respir Crit Care Med. 2015;4:5. [Google Scholar]
- 30*.George BJ, Reif DM, Gallagher JE, et al. Data-driven asthma endotypes defined from blood biomarker and gene expression data. PLoS One. 2015;10(2):e0117445. doi: 10.1371/journal.pone.0117445. A cross-sectional study of 192 predominantly African-American children with asthma from Detroit that, by using a data-driven method, derived several subgroups with distinct mechanisms, such as atopic, mixed eosinophilic and neutrophilic, and TH2-low endotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams-DeVane CR, Reif DM, Hubal EC, et al. Decision tree-based method for integrating gene expression, demographic, and clinical data to determine disease endotypes. BMC Systems Biol. 2013;7:119. doi: 10.1186/1752-0509-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modena BD, Tedrow JR, Milosevic J, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190(12):1363–1372. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinks TS, Brown T, Lau LC, et al. Multidimensional endotyping in patients with severe asthma reveals inflammatory heterogeneity in matrix metalloproteinases and chitinase 3-like protein 1. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comhair SA, McDunn J, Bennett C, Fettig J, Erzurum SC, Kalhan SC. Metabolomic Endotype of Asthma. J Immunol. 2015;195(2):643–650. doi: 10.4049/jimmunol.1500736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lotvall J, Akdis CA, Bacharier LB, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127(2):355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 36.Larkin EK, Hartert TV. Genes associated with RSV lower respiratory tract infection and asthma: the application of genetic epidemiological methods to understand causality. Future Virol. 2015;10(7):883–897. doi: 10.2217/fvl.15.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomsen SF, Stensballe LG, Skytthe A, Kyvik KO, Backer V, Bisgaard H. Increased concordance of severe respiratory syncytial virus infection in identical twins. Pediatrics. 2008;121(3):493–496. doi: 10.1542/peds.2007-1889. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez AE, Marson FA, Bertuzzo CS, Arns CW, Ribeiro JD. Epidemiological and genetic characteristics associated with the severity of acute viral bronchiolitis by respiratory syncytial virus. J Pediatr. 2013;89(6):531–543. doi: 10.1016/j.jped.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 39.Choi EH, Lee HJ, Chanock SJ. Human genetics and respiratory syncytial virus disease: current findings and future approaches. Curr Top Microbiol Immunol. 2013;372:121–137. doi: 10.1007/978-3-642-38919-1_6. [DOI] [PubMed] [Google Scholar]
- 40*.Janssen R, Bont L, Siezen CL, et al. Genetic susceptibility to respiratory syncytial virus bronchiolitis is predominantly associated with innate immune genes. J Infect Dis. 2007;196(6):826–834. doi: 10.1086/520886. The most comprehensive study of RSV infection genetics reporting that the genes from innate immune pathway are associated with the RSV infection severity. [DOI] [PubMed] [Google Scholar]
- 41.Wu P, Larkin EK, Reiss SS, et al. beta2-Adrenergic receptor promoter haplotype influences the severity of acute viral respiratory tract infection during infancy: a prospective cohort study. BMC Med Gene. 2015;16:82. doi: 10.1186/s12881-015-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases: a multiscale approach. J Allergy Clin Immunol. 2015;135(1):31–42. doi: 10.1016/j.jaci.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Mejias A, Dimo B, Suarez NM, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 2013;10(11):e1001549. doi: 10.1371/journal.pmed.1001549. A transcriptomic profiling of whole blood gene expression from U.S. and Finnish children with severe bronchiolitis (either by RSV, rhinovirus, or influenza) that demonstrated pathogen-specific transcriptional signatures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fjaerli HO, Bukholm G, Skjaeret C, Holden M, Nakstad B. Cord blood gene expression in infants hospitalized with respiratory syncytial virus bronchiolitis. J Infect Dis. 2007;196(3):394–404. doi: 10.1086/519168. [DOI] [PubMed] [Google Scholar]
- 45.Fjaerli HO, Bukholm G, Krog A, Skjaeret C, Holden M, Nakstad B. Whole blood gene expression in infants with respiratory syncytial virus bronchiolitis. BMC Infect Dis. 2006;6:175. doi: 10.1186/1471-2334-6-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucasas KL, Mian AI, Demmler-Harrison GJ, et al. Global gene expression profiling in infants with acute respiratory syncytial virus broncholitis demonstrates systemic activation of interferon signaling networks. Pediatr Infect Dis J. 2013;32(2):e68–76. doi: 10.1097/INF.0b013e318278b4b3. [DOI] [PubMed] [Google Scholar]
- 47.van den Kieboom CH, Ahout IM, Zomer A, et al. Nasopharyngeal gene expression, a novel approach to study the course of respiratory syncytial virus infection. Eur Respir J. 2015;45(3):718–725. doi: 10.1183/09031936.00085614. [DOI] [PubMed] [Google Scholar]
- 48.Globinska A, Pawelczyk M, Kowalski ML. MicroRNAs and the immune response to respiratory virus infections. Exp Rev Clin Immunol. 2014;10(7):963–971. doi: 10.1586/1744666X.2014.913482. [DOI] [PubMed] [Google Scholar]
- 49.Leahy TR, McManus R, Doherty DG, et al. Interleukin-15 is associated with disease severity in viral bronchiolitis. Eur Respir J. 2016;47(1):212–222. doi: 10.1183/13993003.00642-2015. [DOI] [PubMed] [Google Scholar]
- 50.Bakre A, Mitchell P, Coleman JK, et al. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J Gen Virol. 2012;93(Pt 11):2346–2356. doi: 10.1099/vir.0.044255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buggele WA, Johnson KE, Horvath CM. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J Biol Chem. 2012;287(37):31027–31040. doi: 10.1074/jbc.M112.387670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalmasso G, Nguyen HT, Yan Y, et al. Microbiota modulate host gene expression via microRNAs. PLoS One. 2011;6(4):e19293. doi: 10.1371/journal.pone.0019293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maudet C, Mano M, Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Letters. 2014;588(22):4140–4147. doi: 10.1016/j.febslet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Collison A, Siegle JS, Hansbro NG, et al. Epigenetic changes associated with disease progression in a mouse model of childhood allergic asthma. Dis Model Mechanism. 2013;6(4):993–1000. doi: 10.1242/dmm.011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathogens. 2007;3(10):1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56**.Moore ML, Stokes KL, Hartert TV. The impact of viral genotype on pathogenesis and disease severity: respiratory syncytial virus and human rhinoviruses. Curr Opin Immunol. 2013;25(6):761–768. doi: 10.1016/j.coi.2013.09.016. A comprehensive review that summarized the impact of respiratory viruses (RSV, rhinovirus) genotypes on the virulence, host immune response, and respiratory infection pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran DN, Pham TM, Ha MT, et al. Molecular epidemiology and disease severity of human respiratory syncytial virus in Vietnam. PLoS One. 2013;8(1):e45436. doi: 10.1371/journal.pone.0045436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinello RA, Chen MD, Weibel C, Kahn JS. Correlation between respiratory syncytial virus genotype and severity of illness. J Infect Dis. 2002;186(6):839–842. doi: 10.1086/342414. [DOI] [PubMed] [Google Scholar]
- 59.Villenave R, Thavagnanam S, Sarlang S, et al. In vitro modeling of respiratory syncytial virus infection of pediatric bronchial epithelium, the primary target of infection in vivo. Proc Natl Acad Sci U S A. 2012;109(13):5040–5045. doi: 10.1073/pnas.1110203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hasegawa K, Linnemann RW, Avadhanula V, et al. Detection of respiratory syncytial virus and rhinovirus in healthy infants. BMC research notes. 2015;8(1):718. doi: 10.1186/s13104-015-1695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller EK, Williams JV, Gebretsadik T, et al. Host and viral factors associated with severity of human rhinovirus-associated infant respiratory tract illness. J Allergy Clin Immunol. 2011;127(4):883–891. doi: 10.1016/j.jaci.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang T, Wang W, Bessaud M, et al. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4(7):e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linsuwanon P, Payungporn S, Samransamruajkit R, et al. High prevalence of human rhinovirus C infection in Thai children with acute lower respiratory tract disease. J Infect. 2009;59(2):115–121. doi: 10.1016/j.jinf.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lau SK, Yip CC, Lin AW, et al. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J Infect Dis. 2009;200(7):1096–1103. doi: 10.1086/605697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65**.Lee WM, Lemanske RF, Jr, Evans MD, et al. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med. 2012;186(9):886–891. doi: 10.1164/rccm.201202-0330OC. An analysis of theChildhood Origins of ASThma (COAST) cohort in Wisconsin that not only found an association of rhinovirus A and C species with severe respiratory infection in infants but also reported a within-species difference in virulence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lynch SV. Viruses and microbiome alterations. Ann Am Thorac Soc. 2014;11(Suppl 1):S57–60. doi: 10.1513/AnnalsATS.201306-158MG. [DOI] [PubMed] [Google Scholar]
- 67.Hyde ER, Petrosino JF, Piedra PA, Camargo CA, Jr, Espinola JA, Mansbach JM. Nasopharyngeal Proteobacteria are associated with viral etiology and acute wheezing in children with severe bronchiolitis. J Allergy Clin Immunol. 2014;133(4):1220–1222. doi: 10.1016/j.jaci.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Mansbach JM, Hasegawa K, Henke DM, et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.01.036. An analysis of the nasopharyngeal microbiota in 1,005 U.S. infants hospitalized for bronchiolitis that found a virus-microbiota associations. The findings were validated in a separate cohort of 307 U.S. children hospitalized for bronchiolitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hasegawa K, Linnemann RW, Mansbach JM, et al. The fecal microbiota profile and bronchiolitis in infants. Pediatrics. 2016 doi: 10.1542/peds.2016-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herbst T, Sichelstiel A, Schar C, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184(2):198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 71.Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336(6080):489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hollams EM, Hales BJ, Bachert C, et al. Th2-associated immunity to bacteria in teenagers and susceptibility to asthma. Eur Respir J. 2010;36(3):509–516. doi: 10.1183/09031936.00184109. [DOI] [PubMed] [Google Scholar]
- 73.Folsgaard NV, Schjorring S, Chawes BL, et al. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am J Respir Crit Care Med. 2013;187(6):589–595. doi: 10.1164/rccm.201207-1297OC. [DOI] [PubMed] [Google Scholar]
- 74.Huang YJ, Nariya S, Harris JM, et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol. 2015;136(4):874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75*.Teo Shu M, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015 doi: 10.1016/j.chom.2015.03.008. A microbiome analysis of nasopharyngeal specimens from Australian infants, that reported that Haemophilus-dominant nasopharyngeal microbiota is associated with a higher incidence of acute respiratory infection and higher severity in infants with high risk of atopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biesbroek G, Tsivtsivadze E, Sanders EA, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190(11):1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 77.Kloepfer KM, Lee WM, Pappas TE, et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. 2014;133(5):1301–7. doi: 10.1016/j.jaci.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carlsson CJ, Vissing NH, Sevelsted A, Johnston SL, Bonnelykke K, Bisgaard H. Duration of wheezy episodes in early childhood is independent of the microbial trigger. J Allergy Clin Immunol. 2015;136(5):1208–1214. doi: 10.1016/j.jaci.2015.05.003. e1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vissing NH, Chawes BL, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med. 2013;188(10):1246–1252. doi: 10.1164/rccm.201302-0215OC. [DOI] [PubMed] [Google Scholar]
- 80.von Linstow ML, Schonning K, Hoegh AM, Sevelsted A, Vissing NH, Bisgaard H. Neonatal airway colonization is associated with troublesome lung symptoms in infants. Am J Respir Crit Care Med. 2013;188(8):1041–1042. doi: 10.1164/rccm.201302-0395LE. [DOI] [PubMed] [Google Scholar]
- 81.Rosas-Salazar C, Shilts MH, Tovchigrechko A, et al. Nasopharyngeal microbiome In RSV resembles profile associated with increased childhood asthma risk. Am J Respir Crit Care Med. 2016 doi: 10.1164/rccm.201512-2350LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herberth G, Offenberg K, Rolle-Kampczyk U, et al. Endogenous metabolites and inflammasome activity in early childhood and links to respiratory diseases. J Allergy Clin Immunol. 2015;136(2):495–497. doi: 10.1016/j.jaci.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 83.Schadt EE, Bjorkegren JL. NEW: network-enabled wisdom in biology, medicine, and health care. Sci Transl Med. 2012;4(115):115rv111. doi: 10.1126/scitranslmed.3002132. [DOI] [PubMed] [Google Scholar]
- 84.Larkin EK, Gebretsadik T, Moore ML, et al. Objectives, design and enrollment results from the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE) BMC Puml Med. 2015;15:45. doi: 10.1186/s12890-015-0040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


