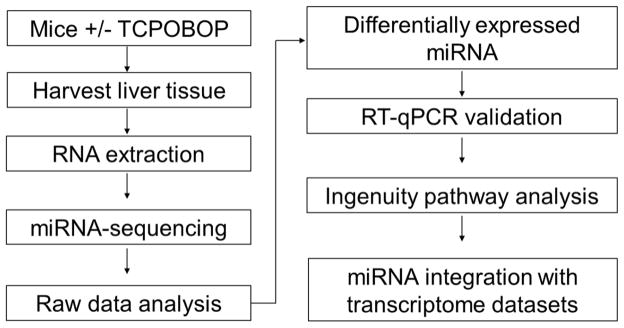
Figure 1.
Scheme of the experimental workflow. Mice were treated with TCPOBOP, and liver tissues were harvested, followed by total RNA extraction. RNA was sequenced on the Illumina HiSeq 2500 platform. Sequencing data were processed using miRDeep2 [29], and differentially expressed miRNAs were determined using DESeq2 package of Bioconductor R. Selected sequencing data were validated using RT-qPCR. The differentially expressed miRNAs were exported into IPA for pathway analysis and integrative analysis with a transcriptome dataset (GSE13688) [45].