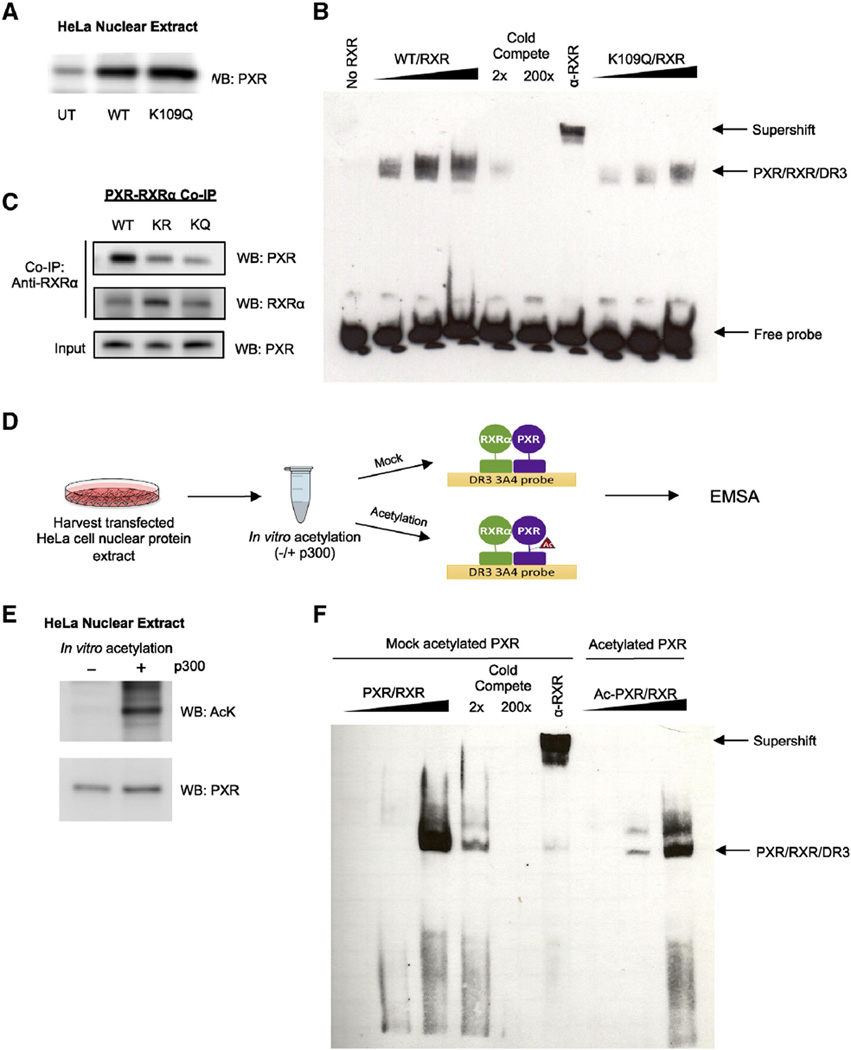
Fig. 6.
Acetylation of PXR inhibits DNA binding and heterodimerization with RXRα. (A) Western blot showing PXR or K109Q ectopic protein expression levels in nuclear extracts harvested from transfected HeLa cells. (B) K109Q mutation reduces DNA binding compared to WT. Increasing amounts of nuclear extract (described in Fig. 5B) enriched with PXR protein (WT vs K109Q) were incubated with nuclear extract enriched with RXRα at a 1:1 ratio along with biotin-labeled oligonucleotide probe containing the DR3 response element. In order to demonstrate specificity, WT reactions were incubated with 2× or 200× unlabeled competing probe or with RXRα antibody. The complexes formed were detected using non-radioactive EMSA. (C) In vitro translated RXRα and WT or mutant PXR were incubated 1:1 at 30 °C for 30 min and then incubated with RXRα antibody-coupled AminoLink Plus Coupling Resin for co-immunoprecipitation. Eluted protein complexes were resolved and analyzed by Western blot to detect amount of PXR that bound to RXRα. (D) Experimental workflow for EMSA using PXR acetylated in vitro. (E) Western blot confirming the acetylation state of the mock acetylated control (−) and acetylated (+) PXR protein samples. (F) In vitro acetylation of PXR reduces DNA binding. PXR-enriched nuclear extracts were mock acetylated or in vitro acetylated (as shown in D) then PXR/RXR/DR3 complex binding was analyzed as described in (B). UT = untransfected, EMSA = electromobility shift assay, IP = immunoprecipitation, WB = Western blot.