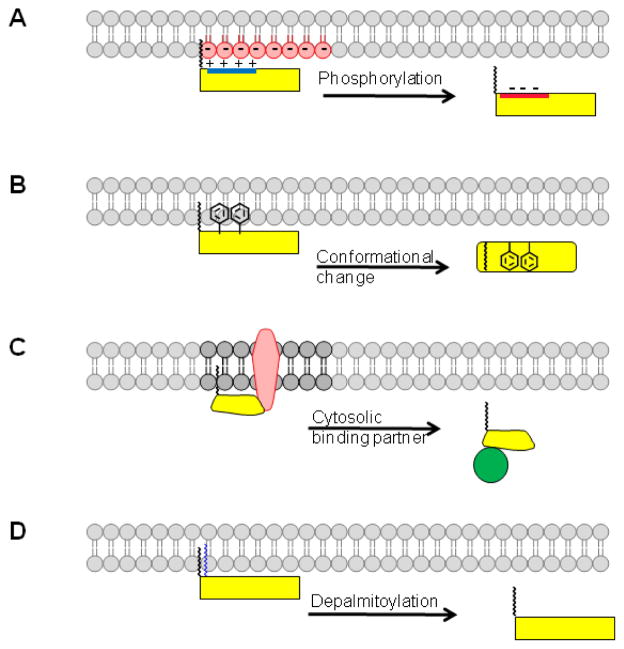
Figure 1. Membrane binding mechanisms for N-myristoylated proteins.
Four different scenarios are illustrated where a second signal, in addition to myristate, contributes to membrane binding. (A) A polybasic cluster of amino acids (blue) enhances membrane binding through electrostatic interactions with head groups of negatively charged phospholipids (red). Phosphorylation of residues within the polybasic cluster reduces or neutralizes the positive charge in the protein, resulting in membrane detachment. (B) Hydrophobic amino acids disposed along the membrane proximal surface of the N-myristoylated protein contribute to membrane binding. Conformational changes can lead to sequestration of the myristate (myristoyl switch) and/or reduced surface accessibility of the hydrophobic residues, leading to release of the protein from the membrane. (C) Interaction with another membrane protein can direct N-myristoylated proteins to the membrane. The protein could detach if another binding partner, with higher affinity, interacts with the N-myristoylated protein and releases it into the cytosol. (D) Palmitoylation of an N-myristoylated protein induces membrane binding, which is reversed upon deacylation.