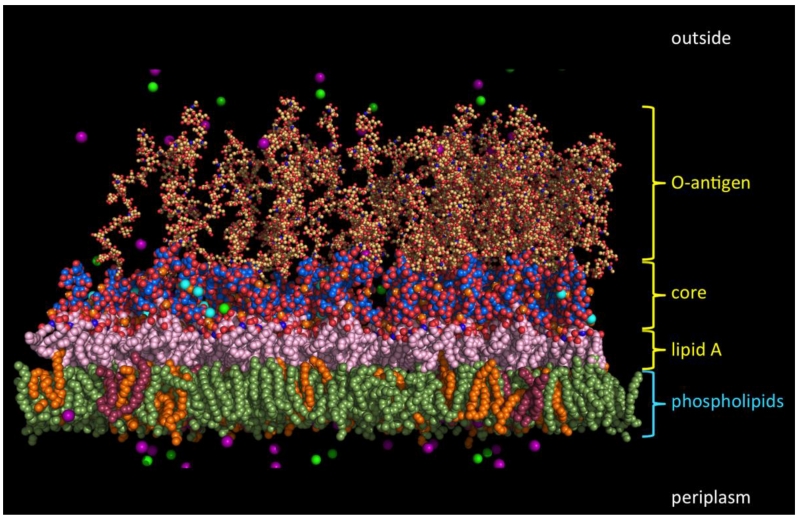
Figure 6. Gram-negative bacterial outer membrane molecular complexity.
The image illustrates a typical E. coli outer membrane and the molecular system used to represent the complexity in molecular dynamics simulations. Molecules represent the bilayer composed of (from the top, external leaflet) glycosylated amphipathic molecules known as lipopolysaccharide consisting of an O-antigen polysaccharide, a core oligosaccharide, and lipid A and (the bottom, periplasmic leaflet) consisting of various phospholipid molecules such as phosphatidylethanolamine (PE; green), phosphatidylglycerol (PG; orange), and cardiolipin (CL; magenta) in a ratio of PE : PG : CL = 15 : 4 : 1. The cyan atoms interspersed with the core oligosaccharides are Ca2+ ions, which immobilize the membrane by mediating the cross-linking electrostatic interaction network with phosphate and carboxyl groups attached to the lipid A and core sugars. Magenta and yellow spheres represent K+ and Cl− ions, respectively.