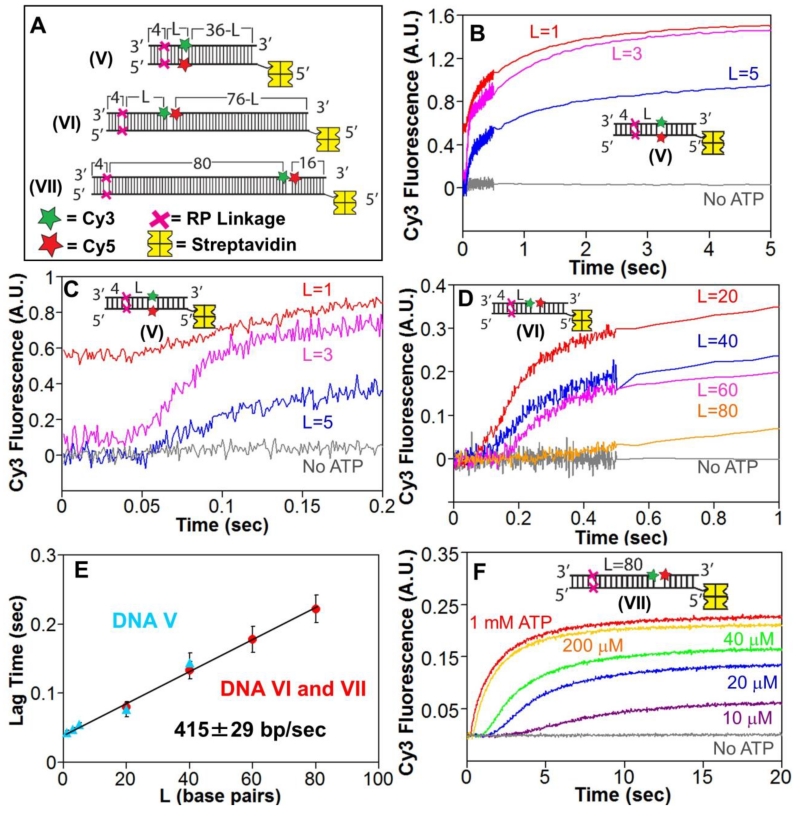
Figure 2. RecBCD can unwind DNA when canonical motor translocation is blocked by RP linkages.
A. DNA used to test if RecBCD can unwind DNA when ssDNA translocation is blocked by RP linkages. A red X indicates the positions of the 3′-3′ and 5′-5′ RP linkages, separated by L bp from a Cy3 (green star) and Cy5 (red star) fluorophores. One end of each DNA is biotinylated so that streptavidin (yellow) can bind and block binding of RecBCD.
B. Stopped-flow time courses were obtained by mixing pre-incubated RecBCD (15 nM) and DNA V (20 nM) with ATP (5 mM) and heparin (8 mg/mL) in Buffer M (30 mM NaCl) at 25° C. Cy3 fluorescence was excited at 505 nm and Cy3 and Cy5 fluorescence were monitored simultaneously. Cy3 fluorescence time courses are shown for DNA V with L=1 bp (red), L=3 bp (pink) and L=5 bp (blue).
C. The first 0.2 seconds of the time courses shown in panel B showing increase in lag time with increasing L.
D. Stopped-flow time courses showing RecBCD unwinding of DNA VI and DNA VII. Experiments were performed as described in panel B. Cy3 time courses for DNA series VI with L=20 bp (red), L=40 bp (blue), L=60 bp (pink) and DNA VII (L=80 bp).
E. Lag times for RecBCD unwinding of RP DNA (5 mM ATP) with Cy3/Cy5 positioned in the DNA backbone (DNA series V) (blue triangles) or across a nick (DNA VI and VII) (red).
F. ATP-dependence of RecBCD unwinding of DNA VII. Experiments were performed as described in panel B, at the indicated [ATP].