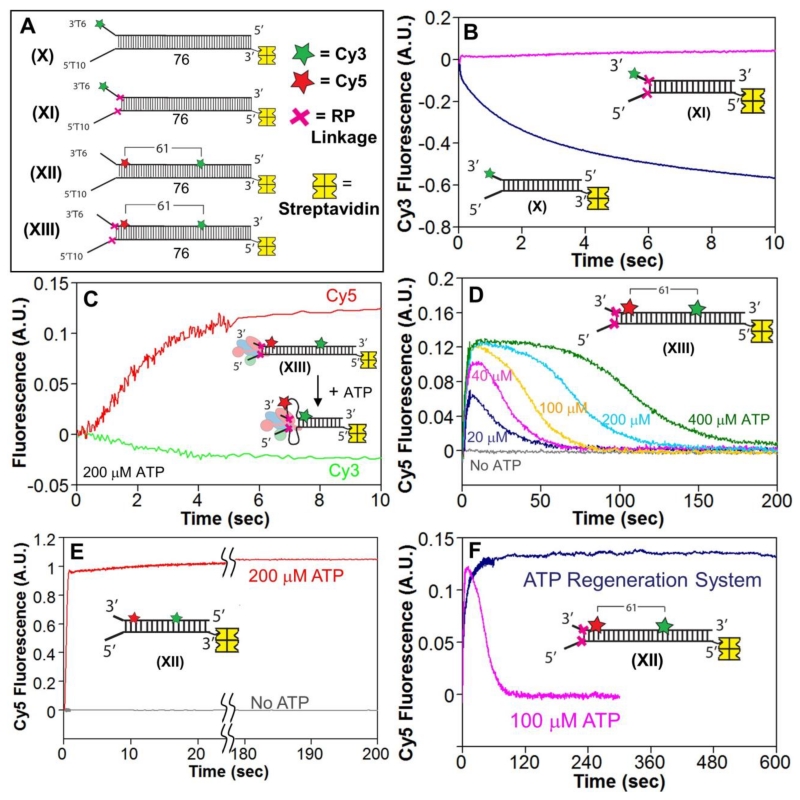
Figure 4. The RecBCD motors remain stuck behind the RP linkages and induce ssDNA loops during DNA unwinding.
A. DNA substrates used to test for ssDNA loop formation during RecBCD unwinding of RP DNA. A red X indicates the positions of the 3′-3′ and 5′-5′ RP linkages. Cy3 (green star) and Cy5 (red star) fluorophores are separated by 61 base pairs. One end of each DNA is biotinylated so that streptavidin (yellow) can bind and block binding of RecBCD.
B. Stopped-flow time courses, monitoring Cy3 fluorescence, for RecBCD unwinding of normal DNA X (blue) and RP DNA XI (pink). RecBCD (18.75 nM) and DNA (25 nM) were pre-incubated and then mixed with ATP (5 mM) and heparin (8 mg/mL) in Buffer M (30 mM NaCl) at 25° C.
C. Stopped-flow time courses showing an increase in FRET signal accompanying RecBD1080ACD unwinding of RP DNA XIII. Experiments were performed by mixing pre-incubated RecBCD (37.5 nM) and DNA XIII (50 nM) with ATP (200 μM) and heparin (8 mg/mL) in Buffer M (30 mM NaCl) at 25° C. Cy3 fluorescence was excited at 505 nm, and Cy3 (green) and Cy5 (red) fluorescence were monitored.
D. Cy3 fluorescence time courses for experiments performed with RecBD1080ACD and RP DNA XIII as described in panel C, at 400 μM ATP (green), 200 μM ATP (light blue), 100 μM ATP (gold), 40 μM ATP (pink), 20 μM ATP (dark blue), and no ATP (gray). Inset-identical stopped-flow experiment performed with RecBD1080ACD and DNA XII at 200 μM ATP.