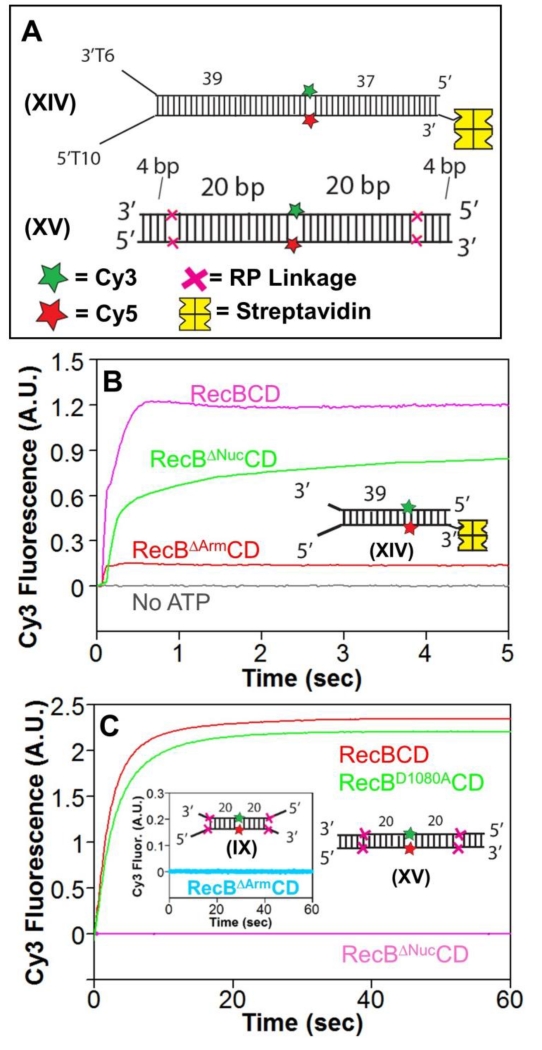
Figure 5. The RecB arm and nuclease domains are needed for RecBCD to unwind RP DNA.
A. DNA substrates used to test for unwinding by RecBΔArmCD and RecBΔNucCD. DNA XIV contains a high affinity binding site on one end and is biotinylated on the opposite end to bind streptavidin (yellow) to block RecBCD binding. Cy3 (green star) and Cy5 (red star) are on opposite strands 39 bp from the binding site. A red X indicates the positions of the 3′-3′ and 5′-5′ RP linkages in DNA XV separated by L bp from a Cy3 (green star) and Cy5 (red star) fluorophores.
B. Stopped-flow Cy3 fluorescence time courses showing unwinding of DNA XIV (20 nM) by RecBCD, RecBΔNucCD, and RecBΔArmCD (15 nM) in Buffer M (30 mM NaCl) (5 mM ATP and 8 mM heparin) at 25° C.
C. Stopped-flow Cy3 fluorescence time courses with RP DNA XV and RecBCD (15 nM) (red), RecBD1080ACD (15 nM) (green), and RecBΔNucCD (15 nM) (pink) in Buffer M30 (5 mM ATP and 8 mg/mL heparin) at 25° C. Inset-RecBΔArmCD is unable to unwind RP DNA IX containing a high affinity binding site under the same conditions.