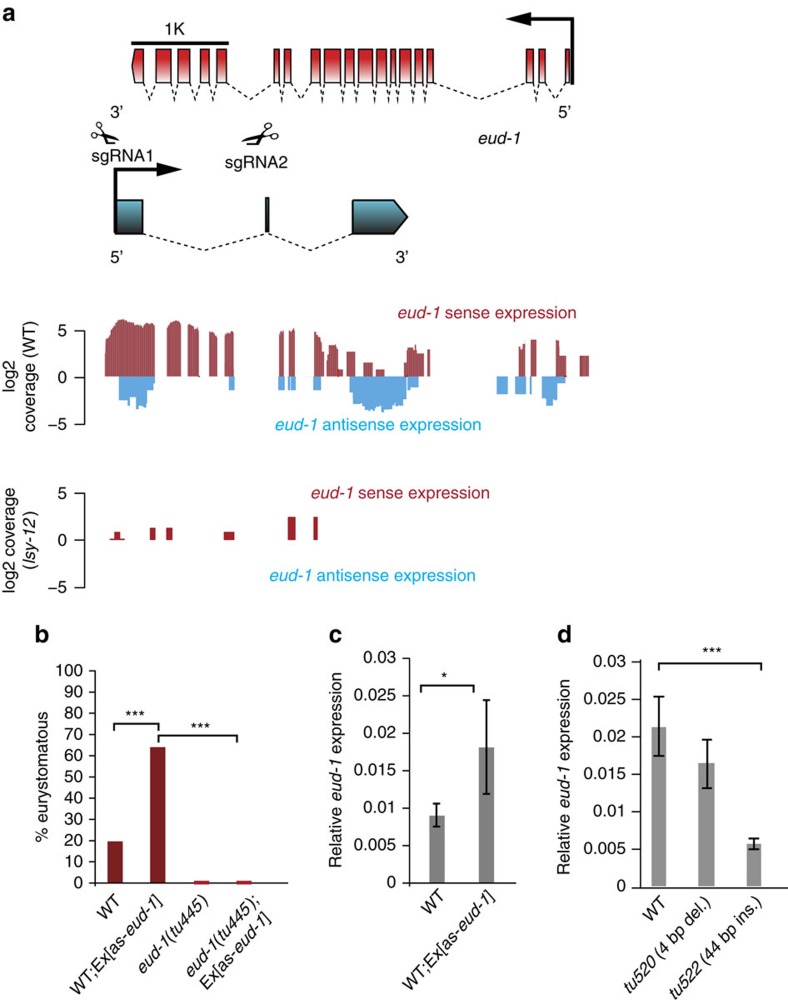
Figure 3. Molecular organization and function of as-eud-1.
(a) Organization of the eud-1 and antisense eud-1 (as-eud-1) locus and RNAseq experiments of wild-type and Ppa-lsy-12 mutant animals. The long noncoding RNA as-eud-1 consists of three exons that span large parts of the eud-1 coding region. The structure of as-eud-1 was identified in RT–PCR experiments and revealed the existence of a short exon, which went undetected in RNAseq. Other antisense reads obtained at lower frequency in the RNAseq experiment, were not confirmed to be part of as-eud-1 in RT–PCR experiments with mixed stage wild-type animals. Subsequent panels show sense and antisense expression as measured for wild-type (wt) and Ppa-lsy-12 mutant animals. Note that nearly no reads of eud-1 and as-eud-1 were observed in Ppa-lsy-12 mutants. sgRNA1 and sgRNA2 in the 5′ untranslated region (UTR) and exon 2 of as-eud-1, respectively, were used to induce mutations by CRISPR. (b) Transformation of wild-type hermaphrodites with as-eud-1 cDNA induced a high incidence of males and a Eud phenotype in male progeny. In contrast, transformation of eud-1(tu445) mutant animals with as-eud-1 did not result in a Eud phenotype, although the high incidence of males was similar to the transformation of wild-type animals. Two independent transgenic lines were generated each, n>100 for all strains. (c) qRT–PCR experiments reveal an up-regulation of eud-1 in as-eud-1 transgenic males relative to wild-type males. This experiment has been replicated three times. Error bars are defined as s.e.m. (d) qRT–PCR experiments reveal that eud-1 is significantly down-regulated in the as-eud-1 promoter mutant tu522 that contains a 44 bp insertion. This experiment has been replicated three times. Error bars are defined as s.e.m. *P<0.05 and ***P<10−5, Kruskal–Wallis test.