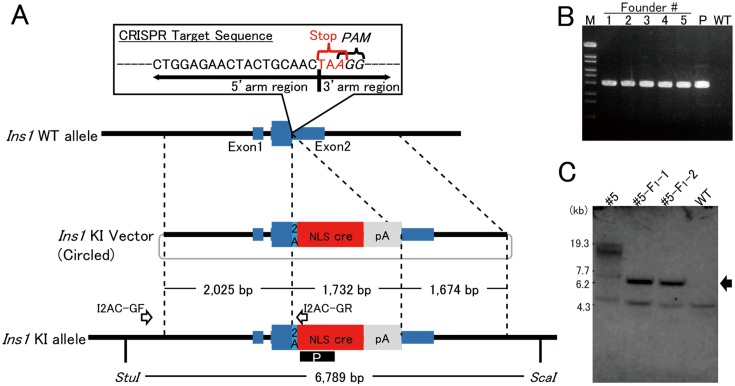
Fig. 1.
Strategy of bicistronic cre expression in pancreatic beta cells. A: To integrate the 2A-cre sequence just before the stop codon of Ins1, the 23-nt sequence (5′-CTGGAGAACTACTGCAACTAAGG-3′) containing both PAM and stop codon was chosen as the CRISPR target. The 3′-end of the region of 5′-homology arm (2.0 kb) is the final coding sequence of Ins1. The 5′-end of the region of 3′-homology arm is the stop codon of Ins1. The arrows labeled I2AC-GF and I2AC-GR indicate the primers for detecting the knock-in allele. The black box including the white letter P indicates the probe used for Southern blotting. B: PCR products, amplified with the founder genome DNA as the template and the primers I2AC-GF and I2AC-GR, of the appropriate size (2,631 bp) were detected. M: Marker 6 (Nippongene). C: Southern blotting analysis with founder #5 and its offspring (#5-F1-1 and #5-F1-2). In #5, the knock-in band (arrow) and longer random integration band were detected. In contrast, the KI band and no random integration band were detected in offspring. Lower weight nonspecific bands were detected in all samples.