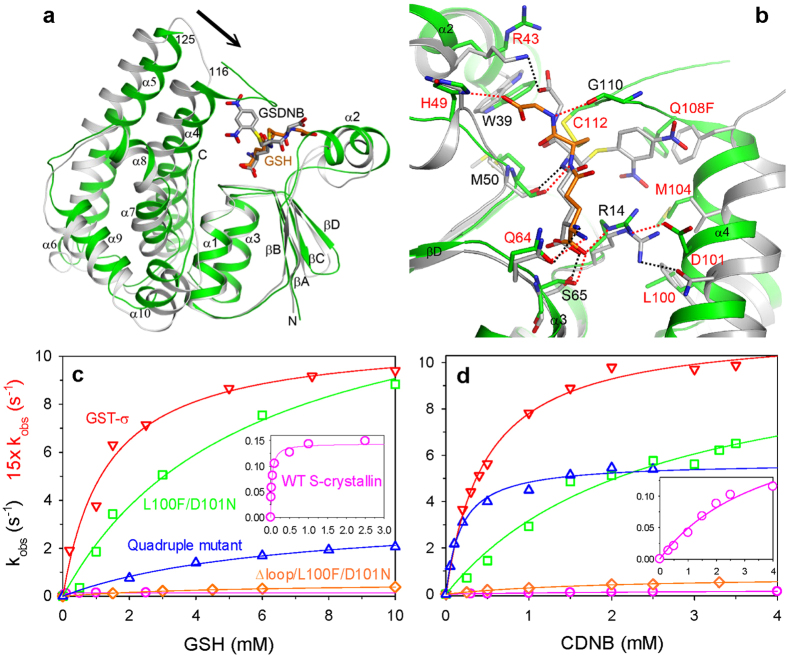
Figure 2. Structural and biochemical characterizations of S-crystallin.
(a) An overlay of the structure of the S-crystallin Q108F mutant in complex with GSH (green) against GST-σ in complex with GSDNB (grey)16. The arrow indicates the conformational differences in the α4 and α5 helices. (b) Overlay of the active sites of the two structures. Residues labeled in red were selected for mutagenesis. (c,d) The GST activity of S-crystallin. Plot of the rate constant as a function of the concentration of two substrates GSH (c) and CDNB (d) for the wild-type (magenta) and various S-crystallin mutants, the L100F/D101N double mutant (green), the ∆loop/L100F/D101N double mutant (orange), and L100F/D101N/M104V/Q108F quadruple mutant (blue). The activity of GST-σ from octopus liver tissues (red) was used as a positive control. Insets show plots of the wild-type S-crystallin at a suitable scale.