Figure 1. Model of Cas9 DNA targeting and cleavage.
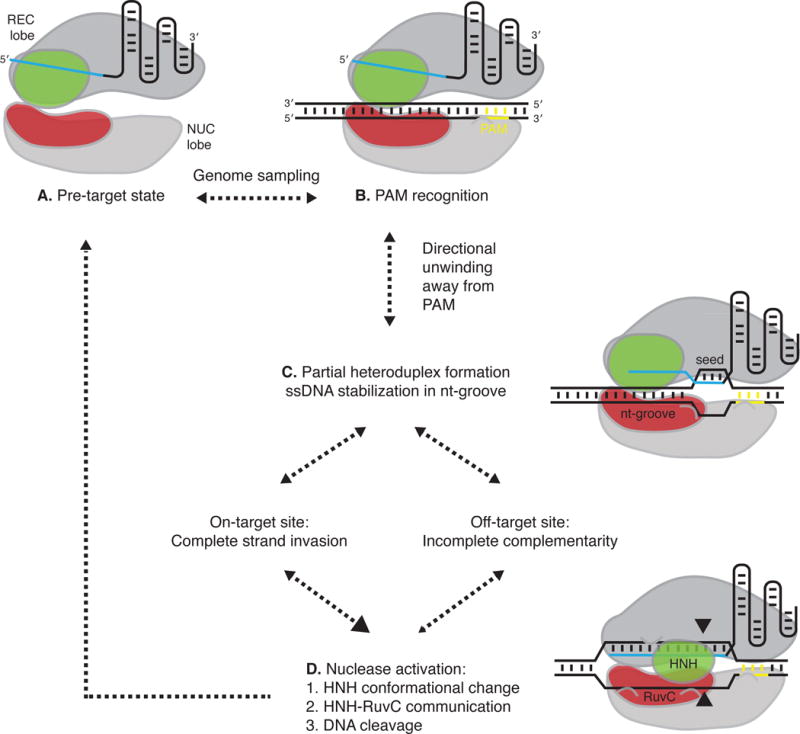
A) Cas9 in complex with guide RNA samples the genome for PAM matches by diffusion, and undersamples heterochromatin regions under certain conditions (Knight et al., 2015). B) Upon PAM recognition and dsDNA binding, the DNA is directionally unwound as the guide RNA interrogates the target strand for complementary sequences. C) The non-target ssDNA strand is stabilized in a non-sequence specific manner in Cas9’s nt-groove. Incomplete complementarity or a short 15-nt PAM-proximal region of complementarity is sufficient for Cas9 binding, as demonstrated by dCas9 or ‘dead-guides’ with wtCas9, respectively. Near-complete complementarity of >17-nt of RNA:DNA heteroduplex is required for nuclease activation. D) After this RNA strand invasion, the HNH domain undergoes a conformational change and communicates through a linker to the RuvC domain. The HNH and RuvC catalytic domains simultaneously cleave the target and non-target ssDNA strand, respectively. Cas9 may remain bound to the cut site for an extended period before returning to the pre-target state (Richardson et al., 2016). The model shown is adapted from several references (Anders et al., 2014; Nishimasu et al., 2014; Sternberg et al., 2014; Sternberg et al., 2015; Jiang et al., 2016; Slaymaker et al., 2016).
