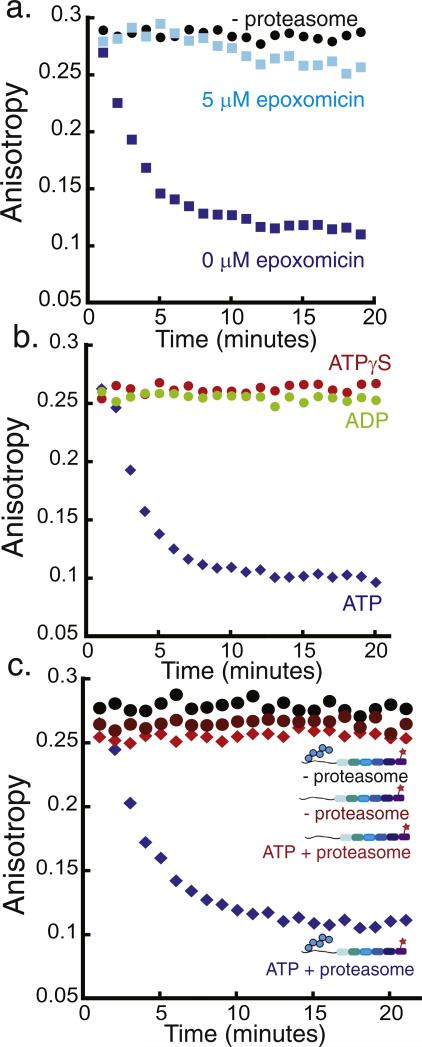
Fig. 3.
Monitoring 26S proteasome activity by fluorescence anisotropy. Degradation of the polyubiquitinated Alexa Fluor 546-labeled substrate by purified yeast proteasome as monitored by fluorescence anisotropy measurements. The degradation reaction was monitored at room temperature over 20 min by performing anisotropy readings every minute. One representative experiment is shown in each graph, and the decay rate constants obtained from the exponential fits to repeat experiments can be found in Supplementary Materials Table S2. The graphs plot fluorescence anisotropy against time. (a) Degradation of approximately 20 nM polyubiquitinated substrate in the presence of 2 mM ATP and 50 nM proteasome (blue squares), 50 nM proteasome + 5 μM epoxomicin (light blue squares), or no proteasome (black circles). (b) Degradation of approximately 20 nM polyubiquitinated substrate by 50 nM proteasome and 2 mM ATP (blue diamonds), 2 mM ADP (green circles), or 2 mM ATPγS (red circles). (c) Degradation of approximately 20 nM polyubiquitinated substrate in the presence of 2 mM ATP and 50 nM proteasome (blue diamonds), degradation of approximately 20 nM substrate lacking the polyubiquitin modification by 50 nM proteasome (red diamonds), and incubation of approximately 20 nM polyubiquitinated substrate without proteasome (black circles).