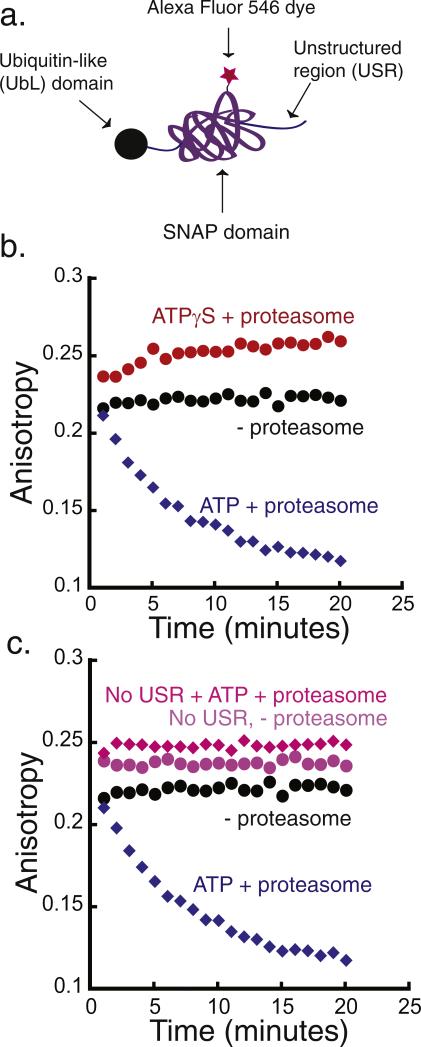
Fig. 4.
Degradation of a simplified model protein. (a) Schematic representation of a model substrate that is targeted to the proteasome by a ubiquitin-like (UbL) domain from S. cerevisiae Rad23 protein. The substrate consists of the UbL domain at its N terminus, followed by the SNAP-tag and a 40-amino acid-long unstructured region at the C terminus that functions as the proteasome initiation site (UbL-SNAP-40-His6). (b) Degradation of UbL-SNAP-40-His6 by purified yeast proteasome monitored by fluorescence anisotropy. The degradation reaction was monitored at room temperature over 20 min, taking an anisotropy reading every minute. Incubation of 20 nM substrate in the presence of 2 mM ATP and 50 nM proteasome (blue diamonds), of 20 nM substrate in the presence of 2 mM ATPγS and 50 nM proteasome (red circles), and of 20 nM substrate alone (black circles). (c) Degradation reaction of UbL substrate with (UbL-SNAP-40-His6) and without (UbL-SNAP-His6) a C-terminal unstructured region. Experiments are as in (b): 20 nM of UbL-SNAP-40-His6 with 50 nM proteasome and 2 mM ATP (blue diamonds), 20 nM UbL-SNAP-His6 with 50 nM proteasome and 2 mM ATP (pink diamonds), 20 nM substrate UbL-SNAP-40-His6 alone (black circles), and 20 nM UbL-SNAP-His6 alone (light purple circles).