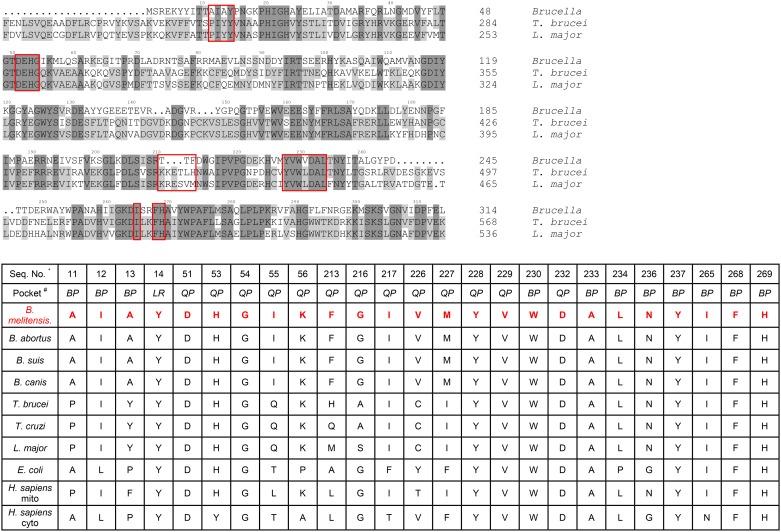
Fig 5. Amino acid sequence alignments within the benzyl and quinolone pocket of BmMetRS, TbMetRS and LmMetRS.
Inhibitor binding residues in BmMetRS compared to TbMetRS and LmMetRS are highlighted in red boxes. The most significant differences can be found in residues 211–213 (TTF) of the BmMetRS which are analogous to a larger run of residues (TbMetRS: KRETLH LmMetRS: KRESVM) in the trypanosome structures. Inhibitor interaction with Phe213 within the BmMetRS complex led to different protein geometry relative to the TbMetRS complex. Table shows list of residues within the benzyl and quinolone pockets interacting with inhibitors relative to other MetRS. Sequence numbers refer to the BmMetRS sequence. # LR = linker region, BP = benzyl pocket, QP = quinolone pocket.