Fig 1.
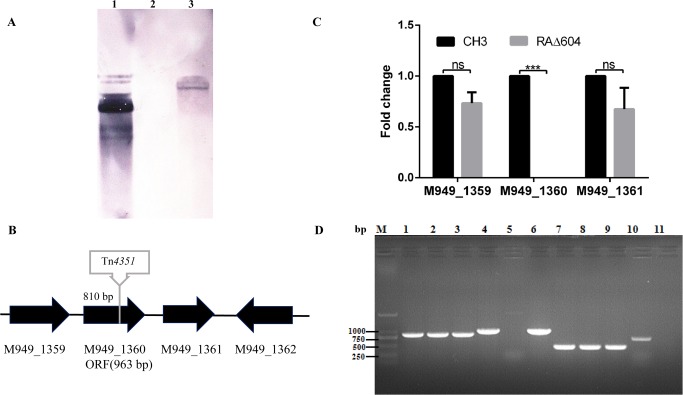
Characterization of R. anatipestifer mutant strain RAΔ604 and complementation strain cRAΔ604 (A) Southern blot analysis. Lane 1: PEP4351 (positive control); Lane 2: R. anatipestifer CH3 (negative control); Lane 3: mutant strain RAΔ604. (B) Schematic chart of Tn4351 insertion in RAΔ604 chromosome at 810 bp of the M949_1360 gene. The flanking genes M949_1359, M949_1361, and M949_1362 were annotated as hypothetical protein, glycosyl transferase group 1 and lipopolysaccharide-modifying protein respectively. (C) qPCR analysis. Changes in mRNA levels are expressed as fold expression and calculated using the comparative CT (2-ΔΔCT) method. The expression of M949_1360 was inactivated in RAΔ604. However, no significant difference was shown in expression levels of M949_1359 and M949_1361 between R. anatipestifer CH3 and RAΔ604 (ns, p > 0.05). Error bars represent standard deviations from three replicates (***, p < 0.01). (D) Identification of the complementation strain cRAΔ604 by PCR. Takara DL2000 marker; lanes 1–3: R. anatipestifer 16S rRNA was amplified from the WT strain CH3 (lane 1), the mutant strain RAΔ604 (lane 2), and the complementation strain cRAΔ604 (lane 3), showing a 744-bp fragment of 16S rRNA; lanes 4–6: 963-bp fragment of M949_1360 gene was amplified from the WT strain CH3 (lane 4), and the complementation strain cRAΔ604 (lane 6), no 963-bp fragment of M949_1360 gene was amplified from the mutant strain RAΔ604 (lane 5); lanes 7–11: a 459-bp fragment of R. anatipestifer dnaB gene, but not a E. coli phoA gene was amplified from the WT strain CH3 (lane 7), the mutant strain RAΔ604 (lane 8) and the complementation strain cRAΔ604 (lane 9); lane 10: a 720-bp fragment of E. coli phoA gene was amplified from E. coli S17-1 strain; lane 11: distilled water, as a negative control.