Figure 6. Wiz binding at the protocadherin β locus in the adult cerebellum.
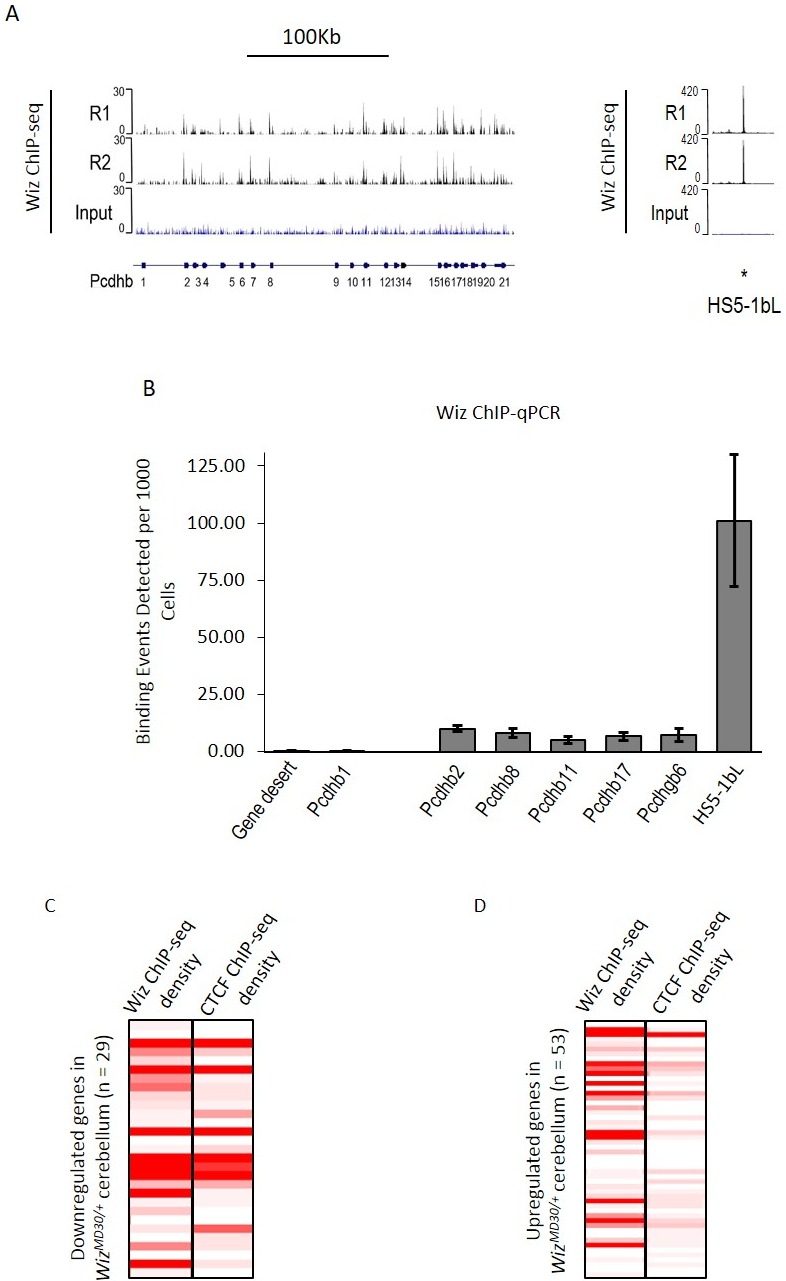
(A) Genome browser screenshots of the protocadherin β locus and the HS5-1bL enhancer (*indicated) located ~400 Kb downstream. Shown are Wiz ChIP-seq data generated in the adult cerebellum as the read depth along the locus. Wiz binds promoters of the Pcdhb locus (left) and is enriched more at the downstream HS5-1bL enhancer (right), the relative binding of Wiz is indicated by the altered scale bar. The location of the Pcdhb genes, are indicated. (B) ChIP-qPCR was performed using the anti-Wiz antibody for two negative Wiz-enrichment sites, with primers located in a gene desert and at Pcdhb1, which was not enriched in ChIP-seq, and six positive Wiz-enrichment sites, including the promoters of four Pcdhb genes, the promoter of a Pcdhg gene and the downstream HS5-1bL enhancer. Enrichment is represented as binding events detected per 1000 cells and are generated by running samples in parallel with known amounts of genomic DNA. Error bars are the SEM from 3 biological replicates. (C) Shown is Wiz ChIP-seq read density in adult cerebellum as a heatplot for the 29 genes that decreased and (D) the 53 genes that increased, in expression in the cerebellum of WizMommeD30/+ mice. Also shown is CTCF ChIP-seq read density. ChIP-seq density was calculated for the 1 Kb surrounding the TSS of each gene and genes are ranked from the lowest to the highest fold change in each case. Datasets were normalized to the size of smallest library.
