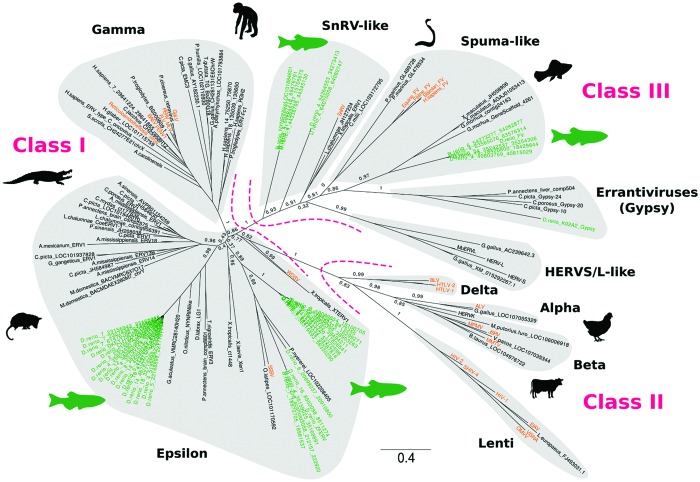
FIGURE 2.
Phylogenetic analysis of zebrafish ERVs and other fish retroelements. The phylogeny was reconstructed from an alignment of RT (201 amino acids, translated from element copies) by Maximum Likelihood using PhyML (Guindon et al., 2010) with optimized parameters (best of NNI and SPR; optimized invariable sites). Branch values represent supporting aLRT non-parametric statistics. Zebrafish and exogenous retrovirus sequences are highlighted in green and orange, respectively. Gypsy LTR retrotransposon sequences were used as an outgroup. The tree was drawn using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).