Table 1. Summary of Data Collection and Refinement Statistics.
| Data Collection (Beamline IO2, Didcot) | |
|---|---|
| Crystal | A140V |
| Parameters | |
| Wavelength (Å) | 0.9795 |
| Resolution (Å)a | 48.52–2.18 (2.30–2.18) |
| Cell bonds (Å) | a = 78.66, b = 53.49, c = 65.78 |
| Cell angles (°) | α = γ = 90°, β = 132.47° |
| Space group | C2 |
| Unique reflections | 10482 (1516) |
| Completeness (%) | 98.9 (98.9) |
| Average redundancy | 3.8 (3.9) |
| Mean I/σ | 9.2 (2.6) |
| Rmerge (%)b | 8.8 (80.3) |
| Refinement | |
| Resolution range of data used (Å) | 48.52–2.18 |
| Reflections used | 40175 (5974) |
| R factor (%)c | 17.5 |
| Free R factor (%)d | 22.7 |
| Average B factors | |
| Wilson B factor (Å2) | 61.8 |
| Protein (Chain A) | 50.8 |
| DNA (Chain B) | 48.9 |
| DNA (Chain C) | 52.3 |
| Water molecules | 69.4 |
| Number of protein molecules in asymmetric unit | 1 |
| Total number of non-hydrogen atoms | |
| Protein | 572 |
| Non-protein | 407 |
| Solvent | 409 |
| RMSD from standard values | |
| Bonds (Å) | 0.0113 |
| Angles (°) | 1.8228 |
| Ramachandran plote | |
| Residues in favoured region (%) | 93.1 |
| Residues in allowed region (%) | 6.9 |
| Residues in disallowed region (%) | 0.0 |
aValues in parentheses are for the highest resolution shell.
bRmerge = 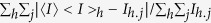 , where
, where 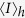 is the mean intensity of symmetry-equivalent reflections.
is the mean intensity of symmetry-equivalent reflections.
cR factor = 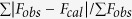 , where Fobs and Fcal are the observed and calculated structure factor amplitudes, respectively.
, where Fobs and Fcal are the observed and calculated structure factor amplitudes, respectively.
dFree R factor value was calculated for R factor using only an unrefined subset of reflections data (5%).
eRamachandran plot was calculated using PROCHECK49.
