Fig. 4.
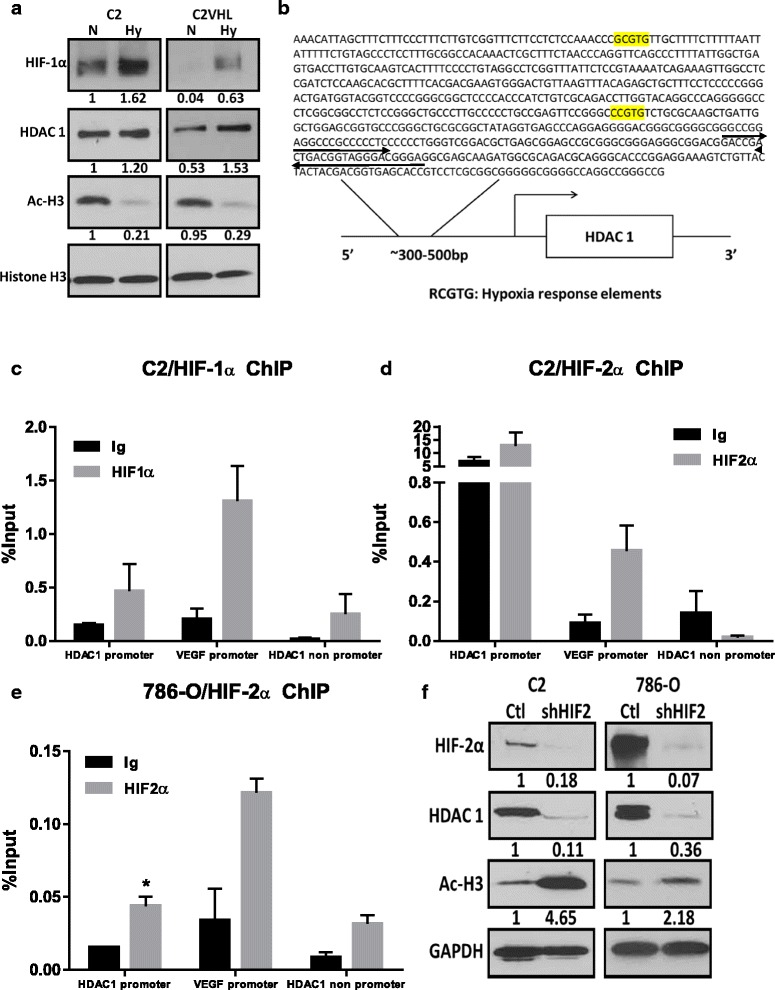
HDAC 1 expression is regulated by HIF in RCC cell lines. a) HDAC 1 expression was compared between C2 and C2VHL cells under normoxic and hypoxic conditions (mimicked by the use of 100 μM cobalt chloride) after overnight serum starvation. The numbers below the bands represent densitometry performed by Image J analysis on representative immunoblots relative to C2 bands in normoxic conditions with total histone H3 serving as loading control. b) HDAC 1 promoter region was analyzed for the presence of hypoxia response elements (HREs) by using the genomatix software. HREs represented by RCGTG and are highlighted in yellow and the black arrow represent the location of the forward and reverse primer. c-e) Chromatin immunoprecipitation assays were carried out in VHL null cell line C2 that expressed both HIF isoforms and 786–0 that only expressed HIF-2α. Pull downs with HIF isoforms were analyzed for HDAC 1 promoter/non-promoter region (1 kb upstream of the transcription start site) and the VEGF promoter (a known target of HIF) by qPCR. The error bars represent standard errors from biological duplicate experiments with technical replicates within each experiment. f) HIF-2α was knocked down in both models of renal cell line tumors by mammalian shRNA and analyzed for HDAC 1 expression and acetylated histone H3 by Western blot. The numbers below the bands represent densitometry performed by Image j analysis on representative immunoblots relative to parental cell lines in normoxic conditions with total GAPDH serving as loading control
