Fig. 4.
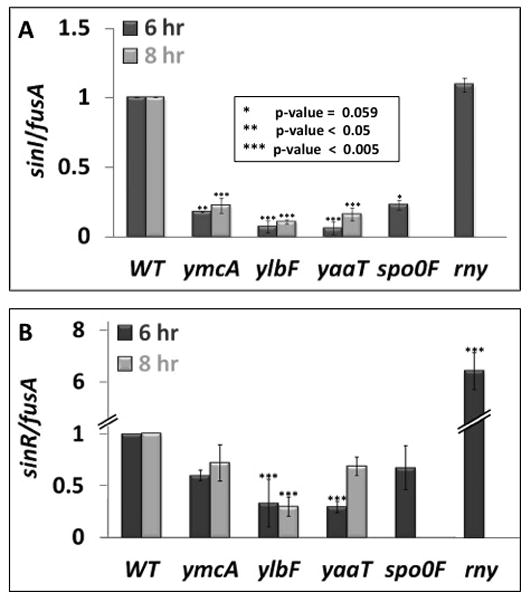
Effects of ylbF, ymcA, yaaT, spo0F and rny knockouts on the abundance of transcripts containing sinI (panel A) and sinR (panel B) sequences. The data were obtained by qRT-PCR and were normalized to the level of fusA mRNA as described in Experimental procedures. Samples for the preparation of RNA were taken from cultures growing in MSgg at the indicated times, which corresponded to late log and stationary phase. The strains used were: wild-type (BD7477), ΔymcA (BD7736), ΔylbF (BD7737), ΔyaaT (BD7731), Δspo0F (BD7593) and Δrny (BD7839). All experiments were repeated 3 independent times, and error bars represent standard deviation. Statistically significant interactions are displayed by asterisks as noted in the figure. Al the strains were constructed in the 3610 background.
