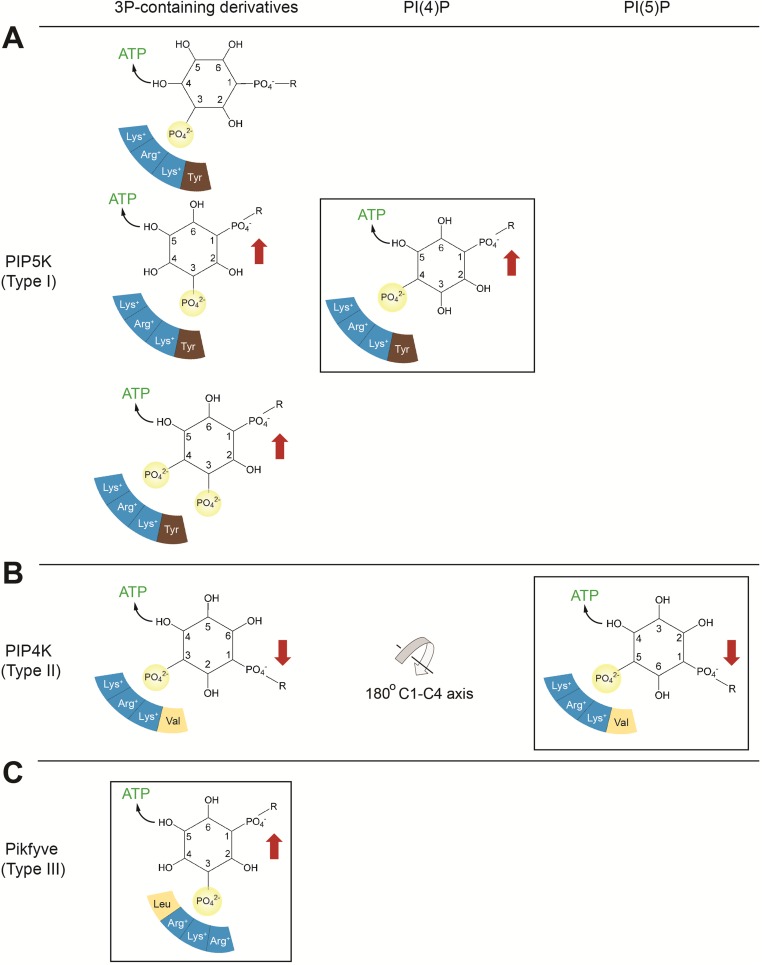
Fig. S5.
Substrate specificity of PIP5K (type I), PIP4K (type II), and PIKfyve (type III). The preferred reaction of each kinase is boxed, and residues of the monophosphate binding site are represented by colored segments, with blue indicating positively charged residues. Red arrows indicate the orientation of the lipid substrate preferred by the specificity loop of each kinase. (A) PIP5K. PIP5K prefers PI(4)P, phosphorylating the 5-hydroxyl, with the 1-phosphate of PI(4)P projecting upward. PIP5K contains a glutamate (Glu382 of PIP5Kα) in the specificity loop, which recognizes the upward orientation of the 1-phosphate. PIP5K cannot phosphorylate the 4-hydroxyl of PI(5)P because this phosphorylation would require the 1-phosphate to project downward. In addition, PIP5K can phosphorylate PI(3)P, generating PI(3,4)P2, PI(3,5)P2, and PI(3,4,5)P3. In the minor reaction that generates PI(3,4)P2, the positions of the 4-hydroxyl and 1-phosphate are not ideal for nucleophilic attack and compatibility with the specificity loop. (B) PIP4K. The alanine (Ala-371 of PIP4Kα) in the specificity loop favors the downward orientation of the 1-phosphate of PI(5)P. A 180° flip around the C1-C4 axis allows the 1-phosphate of PI(3)P to also point downward, compatible with the specificity loop of PIP4K (18). In this orientation, only the 4-hydroxyl can react with ATP. (C) PIKfyve. Compared with PIP5K and PIP4K, PIKfyve features a different arrangement of residues at the phosphate binding site, as shown, causing a slight shift. This shift renders PIKfyve incapable of phosphorylating PI(4)P. The specificity-switch residues of PIKfyve, and thus PIKfyve’s preference for the upward substrate orientation, are identical to those of PIP5K. PIKfyve phosphorylates PI(3)P exclusively at the 5-position.