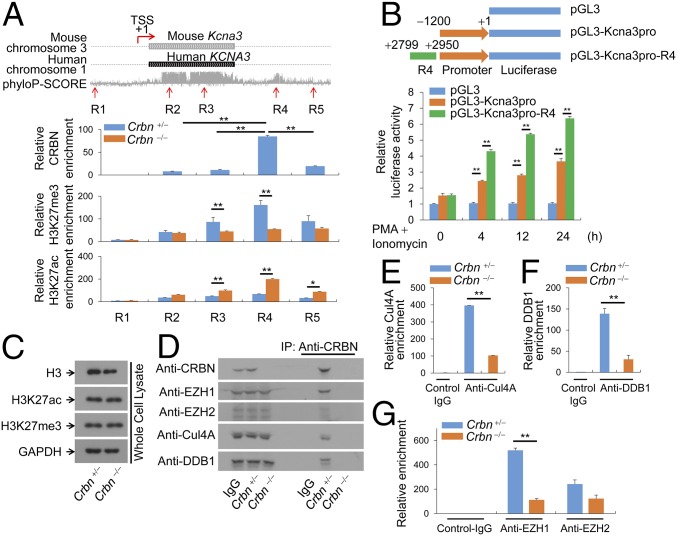
Fig. 3.
CRBN regulates histone modification at Kv1.3 regulatory regions. (A) The Kcna3 region in the mouse and human chromosomes. The phyloP-SCORE shows evolutionary conservation of the bases. TSS, transcription start site. Five regions on mouse Kcna3 are marked as R1, R2, R3, R4, and R5. ChIP was performed with anti-CRBN, anti-H3K27me3, or anti-H3K27ac antibodies, and quantitative PCR analyses for R1–R5 regions were performed. (B) Jurkat T cells were transfected with the indicated reporter plasmid and pRenilla. The cells were then activated with PMA and ionomycin. Renilla luciferase activity served as a reference to normalize gene expression. (C) H3K27me3 and H3K27ac levels in Crbn−/− and Crbn+/− CD4+ T cells were examined by immunoblot analysis with anti-H3K27me3 and anti-H3K27ac antibodies. (D) Binding of CRBN to Cul4A, EZH1, EZH2, or DDB1 in nuclear extracts of CD4+ T cells was analyzed by immunoblot analysis. (E–G) Enrichment of Cul4A (E), DDB1 (F), or EZH1 and EZH2 (G) at Kcna3 was examined by ChIP using anti-Cul4A, anti-DDB1, or anti-EZH1 and anti-EZH2 antibodies. Chromatin was prepared from CRBN-deficient and littermate control CD4+ T cells. After ChIP, DNA fragments were measured by quantitative RT-PCR. Data are representative of two (A) or three (B–G) independent experiments. Results are expressed as mean ± SD. *P < 0.05; **P < 0.01, unpaired two-tailed Student’s t test.