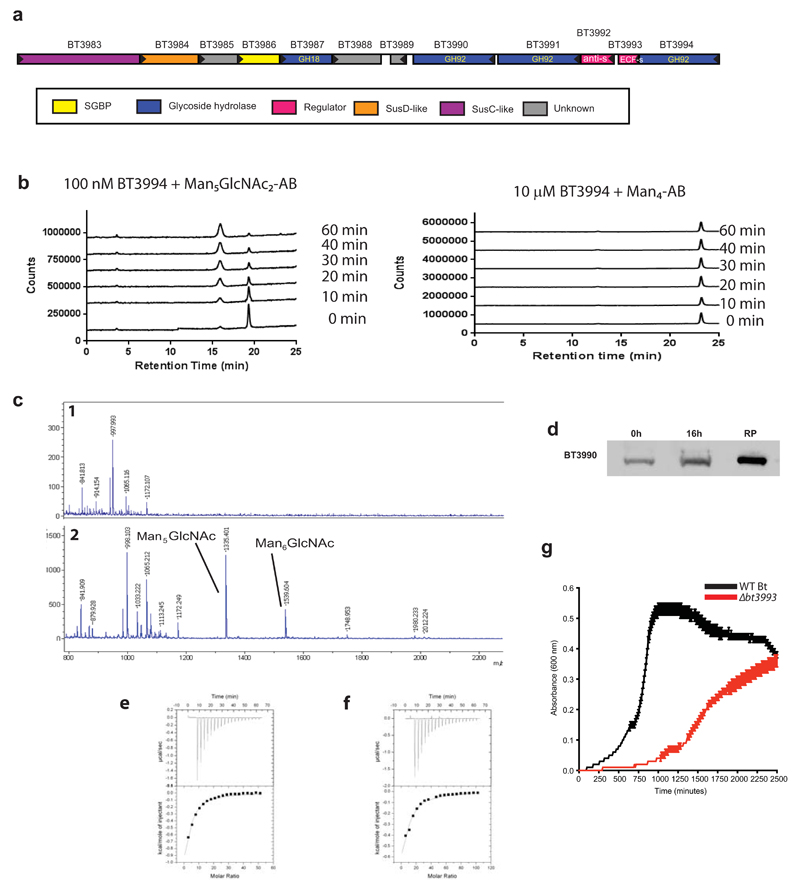
Extended Data Fig. 7. HMNG deconstruction by Bt.
a, Structure of the HMNG PUL. Genes drawn to scale with its orientation indicated. Genes encoding known or predicted functionalities are color-coded and, where appropriate, are also annotated according to their CAZy glycoside hydrolase family (GH) number. SGBP represents a Surface Glycan Binding Protein. b, BT3994 was incubated with α1,6-mannotetraose (Man4-AB) or the high mannose N-glycan Man5GlcNAc2, with both oligosaccharides labelled with 2-aminobenzamide (AB). At the indicated time points aliquots were removed and analysed by HPAEC using a fluorescence detection system. While Man5GlcNac2-AB was hydrolyzed by BT3994, the enzyme was not active against Man4-AB. c, chicken ovalbumin was incubated with buffer (1) or 1 µM of BT3987 (2) in 20 mM Na-HEPES buffer, pH 7.5, for 5 h at 37 °C, and the soluble material was permethylated and analyzed by MALDI-TOF mass spectrometry. The high mannose N-glycans released are labelled. d, depicts a western blot of Bt cells cultured on yeast mannan that were untreated with proteinase K (0 h) or incubated with 2 mg/ml proteinase K for 16 h (16 h). The lane labelled RP contained purified recombinant form of BT3990. The blots were probed with antibodies against BT3990. The data in d are representative of two biological replicates. e, representative isothermal calorimetry titrations (ITCs) for BT3984 titrated with Gal-β1,4-GlcNAc (LacNAc; (25 mM), and f, for BT3986 titrated with mannose (50 mM). The top half of each panel shows the raw ITC heats; the bottom half, the integrated peak areas fitted using a one single binding model by MicroCal Origin software. ITC was carried out in 50 mM Na/HEPES, pH 7.5 at 25 °C. The affinities and thermodynamic parameters of binding are showing in Supplementary Table 5. g, shows the growth profile of wild type Bt (WT Bt; black) and the mutant Δbt3993 (red), which lacks the ECF-σ regulator gene of PUL-HMNG, cultured on Man8GlcNAc2(each curve shows the mean ± s.d. of 3 separate cultures).