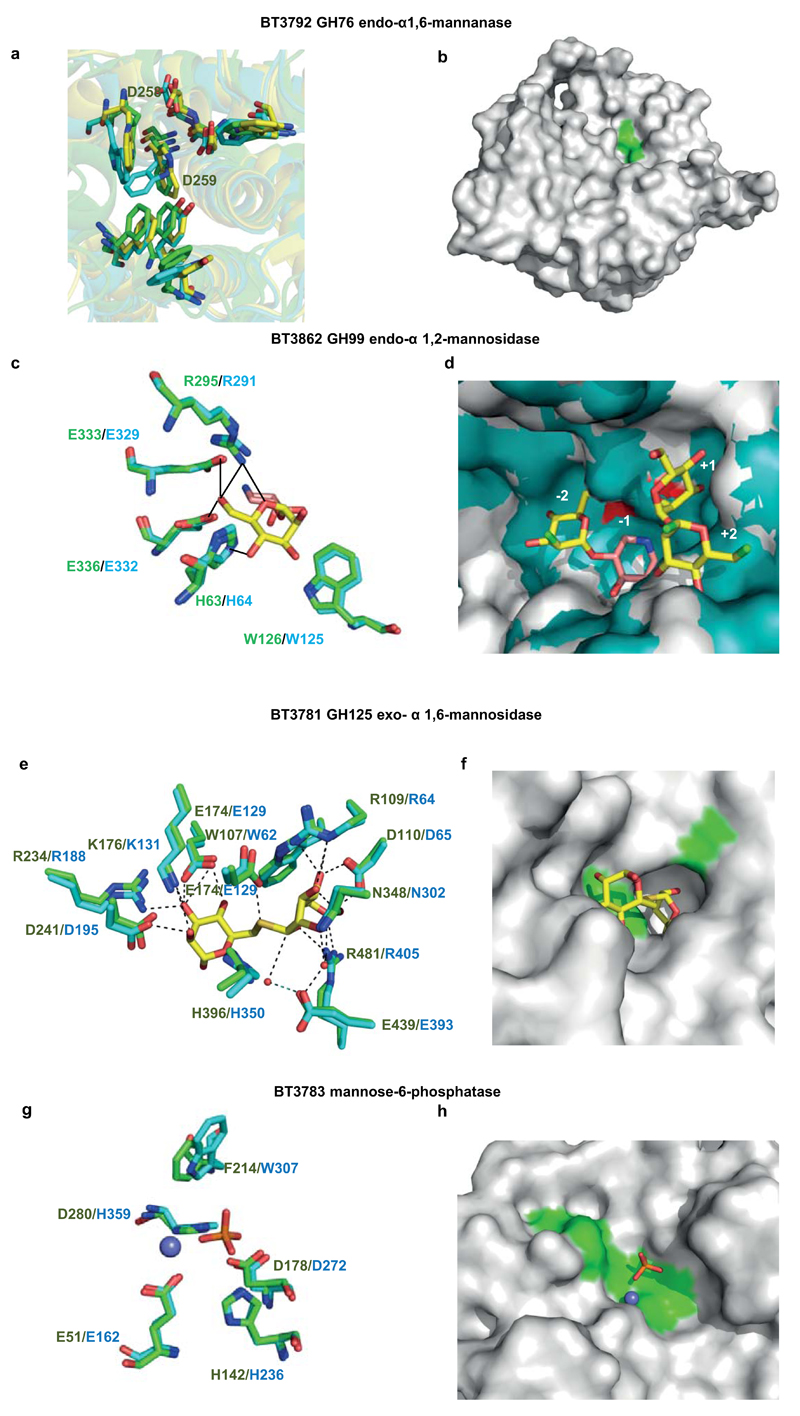
Extended Data Fig. 3. The structures of enzymes that play a key role in yeast mannan degradation.
a, overlay of the hydrophobic conserved residues in the predicted substrate-binding cleft of BT3792 (yellow), BT2949 (cyan) and the Listeria protein Lin0763 (green; PDB code 3K7X), and the predicted catalytic aspartates. b, solvent representation of BT3792 in which the predicted catalytic residues, Asp258 and Asp259, are coloured green. c, overlay of BT3862 (cyan) with a homolog of the enzyme from B. xylanisolvens, BxGH99 (green; PDB code 4UTF) in complex with Man-α1,3-isofagomine and α-1,2-mannobiose (Man residues coloured yellow and isofagomine pink). d, solvent exposed surface of the substrate binding cleft of the BxGH99 (teal) ligand complex overlaid with BT3862 (grey). The subsites are numbered with the catalytic residues, Glu333 and Glu336, coloured red and the solvent exposed O2 of Man bound at the -2 subsite and O1 and O6 of the Man located at the +2 subsite are coloured bright green. e, overlay of BT3781 (green; PDB code 2P0V) with the substrate and catalytic residues of the Clostridium perfringens GH125 α-mannosidase CpGH125 (cyan; PDB code 3QT9), in which the ligand 6-S-α-D-mannopyranosyl-6-thio-a-D-mannopyranose (Man-S-Man) is shown in yellow. f, solvent exposed surface of BT3781 in the vicinity of the active site in which the catalytic residues (Glu174 and Glu439) are depicted in green. The position of Man-S-Man is based on the overlay shown in e. g, overlay of BT3783 with the catalytic and substrate binding residues of a tyrosyl-DNA phosphodiesterase (PDB code 4GYZ) in complex with Mg2+ (slate blue sphere) and phosphate (coloured orange). h, a region of the solvent accessible surface of BT3783 in which the catalytic residues are coloured green. The figure was prepared using PyMOL. A detailed description of the structures of these proteins is provided in Supplementary Information Section 5.