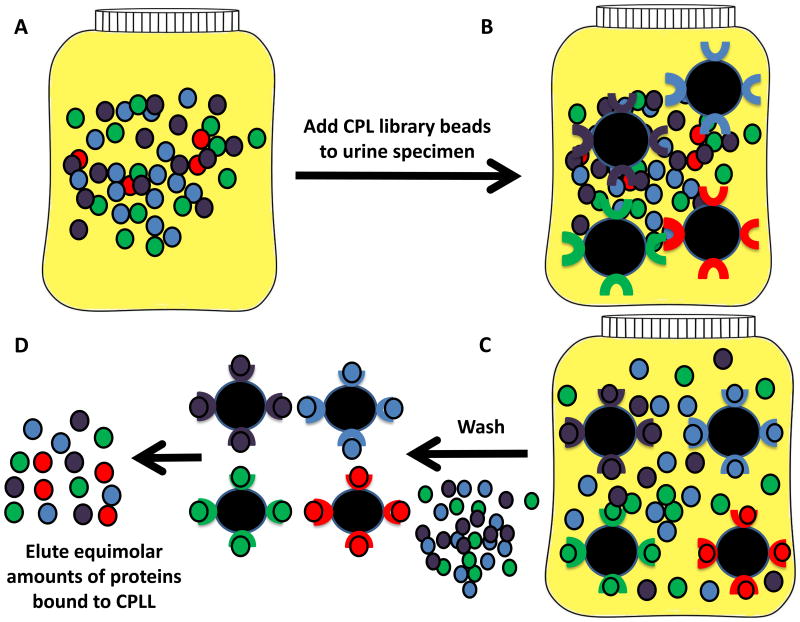
Figure 2. Urine biomarker discovery using combinatorial peptide ligand libraries (CPLL).
CPLL consist of random hexamers of amino acids bound to beads, with one type of hexamer per bead [6, 22, 25, 72]. This approach utilizes a saturation-depletion strategy wherein high abundance proteins will saturate their ligand first, leaving the excess to be washed off, while the low abundance proteins will be able to bind continuously. This strategy minimizes the concentration difference between the low and high abundance proteins. A. Initial urine solution, B. CPLL beads incubated with urine, C. Protein binding to the hexamer bound beads occurs via van der Waals interactions, ionic interaction, and hydrogen bonding. Non-adsorbed, high abundance proteins are removed via washing, D. The hexamer-bound proteins may be recovered en masse, or by protein class, based on standard chromatography elution conditions (pH, chaotropic agent, ionic strength).