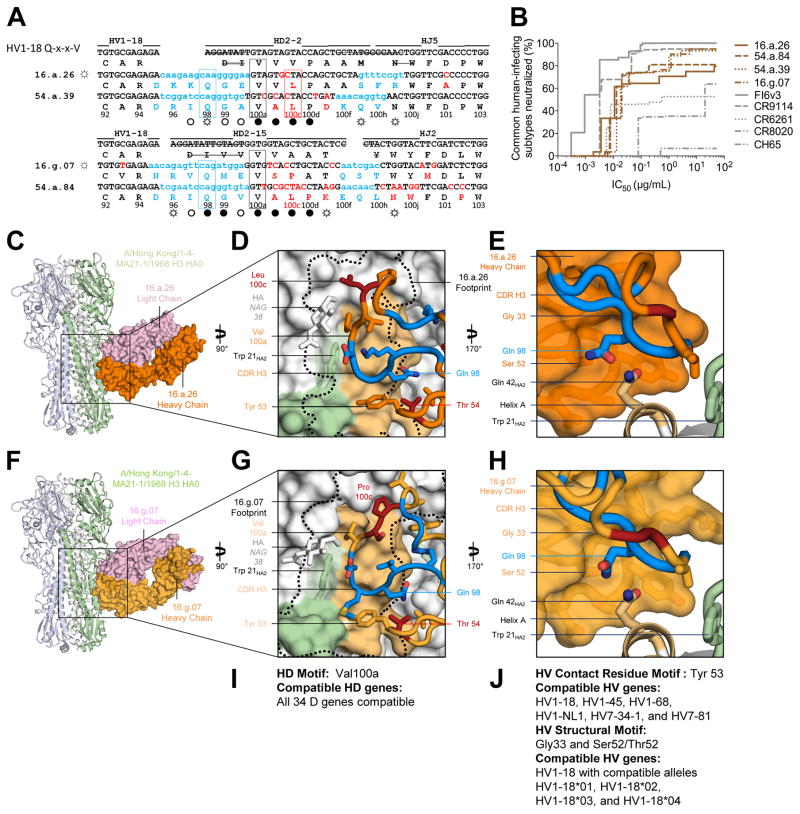
Figure 4. A multidonor HV1-18 class of broadly neutralizing antibodies with Q-x-x-V motif.
(A) Immunoglobulin heavy chains utilizing germline genes HV1-18, HD2-2 or HD2-15 and HJ5 or HJ2, with sequences annotated as described in Figure 2A. (B) Neutralization breadth-potency curve for HV1-18 (Q-x-x-V) class antibodies on a panel of influenza A viruses that includes subtypes known to infect humans. (C) Co-crystal structure of Fab 16.a.26 (HV1-18, HD2-2, HJ5) in complex with H3-HK68. Fab heavy and light chains colored orange and lavender respectively and depicted in surface representation while HA is depicted in ribbon and shown as a trimer. (D) The 16.a.26 CDR H3 is depicted with junction-encoded and mutated residues colored as in panel (A) and germline-encoded residues in orange with the antibody footprint on the HA outlined in black. (E) Antibody heavy chain depicted in surface representation with CDR H3 loop in ribbon. 16.a.26 Gln98HC occupies a turn in the CDR H3, which interacts with the conserved residue Gln42HA2. (F) Co-crystal structure of Fab 16.g.07 (HV1-18, HD2-15, and HJ2) in complex with H3-HK68 depicted as in panel (C). (G) The 16.g.07 CDR H3 depicted as in panel (D) with the antibody footprint on the HA outlined in black. (H) The 16.g.07 CDR H3 is depicted as in panel (E) with Gln98HC contacting Gln42HA2. (I) Analysis of D-gene compatibility. (J) Analysis of V-gene compatibility. See also Figures S3–S7 and Tables S3–S7.